Gene
KWMTBOMO13984 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011337
Annotation
PREDICTED:_regucalcin-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 3.22
Sequence
CDS
ATGTCGGTGAGAATTGAAAAAATCACGGAGCCCCTAACACTGGGGGAAGGGCCCCACTGGGATGAACGTCAGCAAGCGCTATACTTTGTGAGCATCCAAGATAAAACTATACACAAATATGTACCAACTACTGAAAAACATACAAAAACCAGTTTAGATGGGAGGGTTGGTTTCATATTACCAGTGGAAGGTACAACAGACCAATTTGTAGTGGGAGTCGAACGCAAGTTCCTTTTCATACAGTGGGATGGAGAGGATGGCAGTAAAGTTGCTGTATTAAAGGAATTAGGAGAAGTAGACAAAGACAGACCTAATAACAGGATTAATGATGGCAAAGCAGATCCTCGTGGGAGGTTGTTTGCTGGAACTATGGGTCATGAAGATCCTCCAGGTAACTTTGAGAGAAACAAAGCCTCTCTTTACAAATTGGATTCAGCTAAAGATGGGAAACTAGAGAAGATTATCGAAACTGTGTCATTATCAAACGGCCTTGCTTGGGACTTGAAGGAAAAGGCATTTTATTACACAGATTCCATGCAGTTTTCGATAACGAAATTTGATTATGATGTAGATACTGGAGAAATATCCAACCCCAGAAACATATTTGACTTTAAACAGCGCGGTCTACAAGGCATTCCGGACGGTACAACCATCGACACCGACGGCAACCTTTGGGTGGCCGTGTTTGGAGGCTCATGCGTGCTCAAGATCAATCCTAAAAACGGAGAAATCCTACAAAAATTAGCAATTCCCGCCGAACAAGTAACTTCGGCGACTTTCGGGGGCCCAAATCTGGATATTTTATTTGTAACTTCTGCTTGTTGTAATGTTGGTAAGGAGCAGCTCCCACCCTCCGGAGCTACTTTTATGGTCACCGGGCTGGGAGTGAAAGGGCTTCCCAATGTCAGTGTCAGATTGTGA
Protein
MSVRIEKITEPLTLGEGPHWDERQQALYFVSIQDKTIHKYVPTTEKHTKTSLDGRVGFILPVEGTTDQFVVGVERKFLFIQWDGEDGSKVAVLKELGEVDKDRPNNRINDGKADPRGRLFAGTMGHEDPPGNFERNKASLYKLDSAKDGKLEKIIETVSLSNGLAWDLKEKAFYYTDSMQFSITKFDYDVDTGEISNPRNIFDFKQRGLQGIPDGTTIDTDGNLWVAVFGGSCVLKINPKNGEILQKLAIPAEQVTSATFGGPNLDILFVTSACCNVGKEQLPPSGATFMVTGLGVKGLPNVSVRL
Summary
Uniprot
H9JP83
A0A2H1WUP7
I4DP01
I4DLC0
A0A2A4JIX8
A0A0L7L6J1
+ More
A0A0N0PBN4 A0A212FHT2 A0A194QJM9 A0A0N1I833 I4DLB5 A0A194RFD5 A0A2A4JI81 H9JP17 A0A194QNP8 A0A194QNQ3 A0A212FHS6 A0A3S2LTM3 A0A2Z5U7W2 E0XEM8 A0A1Y1KUS1 A0A2J7PV34 D6WJC9 J3JZF0 A0A0L7L6D7 A0A2J7PV36 A0A0N1IES4 K7J5D9 A0A194QHX1 M9NHL6 A0A194QI87 U4TVX1 U5EWT8 A0A139WHG8 A0A194QHS8 A0A336KBK1 A0A1L8DLY9 A0A336KH96 A0A1B6DWB3 A0A1W4XKZ3 A0A1L8DM86 A0A0Q9X2K6 M9NK07 A0A1W6EW18 A0A2A3E259 A0A087ZSJ2 A0A1I8PKJ7 A0A194QJM0 A0A1W4XLV1 A0A0N1IP04 F4W8D2 A0A2P8Z9M7 V5I8P9 A0A0M8ZQU0 A0A0L7RA20 M4QVG7 M4QKU5 M4QII6 A0A158NKR2 B4MG80 A0A1B6G0U5 D6WJC8 Q9NDP1 A0A151IV06 A0A1B6GG13 A0A195BA50 A0A1B0DL94 A0A0Q9VYG4 A0A1D2NDD8 A0A1W6EWA9 M4QIK2 A0A1B6JPW2 R4G322 A0A1B6J451 M4QID0 T1HLA6 A0A182GDM5 Q9BID6 T1PDE1 Q16IH7 A0A1I8N509 A0A0J7KWI6 J9JXU7 A0A1B6LND9 A0A084VYL7 A0A1W4XBG2 A0A0C9PVI0 A0A1W4XBP4 A0A1Q3FSM1 D6WJC6 A0A2H8TR03 A0A0C9REN7 A0A1I8N523 A0A1W4XBF7 E9IDS8 J9LYB7 W8ANX0 T1HLF0 A0A0P8XYJ7 A0A1J1HLE1
A0A0N0PBN4 A0A212FHT2 A0A194QJM9 A0A0N1I833 I4DLB5 A0A194RFD5 A0A2A4JI81 H9JP17 A0A194QNP8 A0A194QNQ3 A0A212FHS6 A0A3S2LTM3 A0A2Z5U7W2 E0XEM8 A0A1Y1KUS1 A0A2J7PV34 D6WJC9 J3JZF0 A0A0L7L6D7 A0A2J7PV36 A0A0N1IES4 K7J5D9 A0A194QHX1 M9NHL6 A0A194QI87 U4TVX1 U5EWT8 A0A139WHG8 A0A194QHS8 A0A336KBK1 A0A1L8DLY9 A0A336KH96 A0A1B6DWB3 A0A1W4XKZ3 A0A1L8DM86 A0A0Q9X2K6 M9NK07 A0A1W6EW18 A0A2A3E259 A0A087ZSJ2 A0A1I8PKJ7 A0A194QJM0 A0A1W4XLV1 A0A0N1IP04 F4W8D2 A0A2P8Z9M7 V5I8P9 A0A0M8ZQU0 A0A0L7RA20 M4QVG7 M4QKU5 M4QII6 A0A158NKR2 B4MG80 A0A1B6G0U5 D6WJC8 Q9NDP1 A0A151IV06 A0A1B6GG13 A0A195BA50 A0A1B0DL94 A0A0Q9VYG4 A0A1D2NDD8 A0A1W6EWA9 M4QIK2 A0A1B6JPW2 R4G322 A0A1B6J451 M4QID0 T1HLA6 A0A182GDM5 Q9BID6 T1PDE1 Q16IH7 A0A1I8N509 A0A0J7KWI6 J9JXU7 A0A1B6LND9 A0A084VYL7 A0A1W4XBG2 A0A0C9PVI0 A0A1W4XBP4 A0A1Q3FSM1 D6WJC6 A0A2H8TR03 A0A0C9REN7 A0A1I8N523 A0A1W4XBF7 E9IDS8 J9LYB7 W8ANX0 T1HLF0 A0A0P8XYJ7 A0A1J1HLE1
Pubmed
EMBL
BABH01009637
ODYU01011140
SOQ56696.1
AK403222
BAM19641.1
AK402088
+ More
BAM18710.1 NWSH01001330 PCG71668.1 JTDY01002624 KOB71072.1 KQ460880 KPJ11518.1 AGBW02008466 OWR53293.1 KQ458761 KPJ05120.1 KPJ11522.1 AK402083 BAM18705.1 KQ460297 KPJ16169.1 PCG71671.1 KPJ05106.1 KPJ05111.1 OWR53292.1 RSAL01000241 RVE43645.1 AP017497 BBB06781.1 GU013474 ADK55065.1 GEZM01075834 GEZM01075833 JAV63930.1 NEVH01020989 PNF20197.1 KQ971343 EFA03133.1 BT128631 AEE63588.1 KOB71073.1 PNF20199.1 KPJ11521.1 AAZX01008996 KPJ05107.1 JN036578 AFJ24877.1 KPJ05109.1 KB631643 ERL84922.1 GANO01000405 JAB59466.1 KYB27438.1 KPJ05108.1 UFQS01000275 UFQT01000275 SSX02284.1 SSX22660.1 GFDF01006606 JAV07478.1 SSX02283.1 SSX22659.1 GEDC01007379 JAS29919.1 GFDF01006607 JAV07477.1 CH964232 KRF99329.1 JN036579 AFJ24878.1 KY563502 ARK19911.1 KZ288446 PBC25614.1 KPJ05110.1 KPJ11519.1 GL887908 EGI69423.1 PYGN01000136 PSN53206.1 GALX01004305 JAB64161.1 KQ435903 KOX69067.1 KQ414619 KOC67710.1 KC689140 AGH29386.1 KC689141 AGH29387.1 KC689138 KC689139 KC689142 KC689143 KC689145 KC689146 KC689147 KC689148 KC689149 KC689150 KC689151 KC689152 KC689154 KC689155 KC689156 AGH29384.1 AGH29385.1 AGH29388.1 AGH29389.1 AGH29391.1 AGH29392.1 AGH29393.1 AGH29394.1 AGH29395.1 AGH29396.1 AGH29397.1 AGH29398.1 AGH29400.1 AGH29401.1 AGH29402.1 ADTU01018655 CH940672 EDW57403.2 GECZ01013723 JAS56046.1 EFA03134.2 AB036903 BAA99282.1 KQ980926 KYN11376.1 GECZ01008401 JAS61368.1 KQ976537 KYM81413.1 AJVK01016346 AJVK01016347 KRF77757.1 LJIJ01000079 ODN03278.1 KY563576 ARK19985.1 KC689153 AGH29399.1 GECU01006486 JAT01221.1 GAHY01002071 JAA75439.1 GECU01013752 JAS93954.1 KC689144 AGH29390.1 ACPB03010982 JXUM01056449 KQ561910 KXJ77200.1 AY028616 AAK26174.1 KA646140 AFP60769.1 CH478076 EAT34066.1 LBMM01002383 KMQ94897.1 ABLF02026048 GEBQ01028432 GEBQ01014707 GEBQ01009166 GEBQ01008064 JAT11545.1 JAT25270.1 JAT30811.1 JAT31913.1 ATLV01018378 KE525231 KFB43061.1 GBYB01005453 JAG75220.1 GFDL01004480 JAV30565.1 EFA03136.1 GFXV01004227 MBW16032.1 GBYB01005451 JAG75218.1 GL762535 EFZ21282.1 ABLF02034011 GAMC01020362 GAMC01020361 JAB86193.1 CH902617 KPU79867.1 CVRI01000006 CRK88362.1
BAM18710.1 NWSH01001330 PCG71668.1 JTDY01002624 KOB71072.1 KQ460880 KPJ11518.1 AGBW02008466 OWR53293.1 KQ458761 KPJ05120.1 KPJ11522.1 AK402083 BAM18705.1 KQ460297 KPJ16169.1 PCG71671.1 KPJ05106.1 KPJ05111.1 OWR53292.1 RSAL01000241 RVE43645.1 AP017497 BBB06781.1 GU013474 ADK55065.1 GEZM01075834 GEZM01075833 JAV63930.1 NEVH01020989 PNF20197.1 KQ971343 EFA03133.1 BT128631 AEE63588.1 KOB71073.1 PNF20199.1 KPJ11521.1 AAZX01008996 KPJ05107.1 JN036578 AFJ24877.1 KPJ05109.1 KB631643 ERL84922.1 GANO01000405 JAB59466.1 KYB27438.1 KPJ05108.1 UFQS01000275 UFQT01000275 SSX02284.1 SSX22660.1 GFDF01006606 JAV07478.1 SSX02283.1 SSX22659.1 GEDC01007379 JAS29919.1 GFDF01006607 JAV07477.1 CH964232 KRF99329.1 JN036579 AFJ24878.1 KY563502 ARK19911.1 KZ288446 PBC25614.1 KPJ05110.1 KPJ11519.1 GL887908 EGI69423.1 PYGN01000136 PSN53206.1 GALX01004305 JAB64161.1 KQ435903 KOX69067.1 KQ414619 KOC67710.1 KC689140 AGH29386.1 KC689141 AGH29387.1 KC689138 KC689139 KC689142 KC689143 KC689145 KC689146 KC689147 KC689148 KC689149 KC689150 KC689151 KC689152 KC689154 KC689155 KC689156 AGH29384.1 AGH29385.1 AGH29388.1 AGH29389.1 AGH29391.1 AGH29392.1 AGH29393.1 AGH29394.1 AGH29395.1 AGH29396.1 AGH29397.1 AGH29398.1 AGH29400.1 AGH29401.1 AGH29402.1 ADTU01018655 CH940672 EDW57403.2 GECZ01013723 JAS56046.1 EFA03134.2 AB036903 BAA99282.1 KQ980926 KYN11376.1 GECZ01008401 JAS61368.1 KQ976537 KYM81413.1 AJVK01016346 AJVK01016347 KRF77757.1 LJIJ01000079 ODN03278.1 KY563576 ARK19985.1 KC689153 AGH29399.1 GECU01006486 JAT01221.1 GAHY01002071 JAA75439.1 GECU01013752 JAS93954.1 KC689144 AGH29390.1 ACPB03010982 JXUM01056449 KQ561910 KXJ77200.1 AY028616 AAK26174.1 KA646140 AFP60769.1 CH478076 EAT34066.1 LBMM01002383 KMQ94897.1 ABLF02026048 GEBQ01028432 GEBQ01014707 GEBQ01009166 GEBQ01008064 JAT11545.1 JAT25270.1 JAT30811.1 JAT31913.1 ATLV01018378 KE525231 KFB43061.1 GBYB01005453 JAG75220.1 GFDL01004480 JAV30565.1 EFA03136.1 GFXV01004227 MBW16032.1 GBYB01005451 JAG75218.1 GL762535 EFZ21282.1 ABLF02034011 GAMC01020362 GAMC01020361 JAB86193.1 CH902617 KPU79867.1 CVRI01000006 CRK88362.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000007151
UP000053268
+ More
UP000283053 UP000235965 UP000007266 UP000002358 UP000030742 UP000192223 UP000007798 UP000242457 UP000005203 UP000095300 UP000007755 UP000245037 UP000053105 UP000053825 UP000005205 UP000008792 UP000078492 UP000078540 UP000092462 UP000094527 UP000015103 UP000069940 UP000249989 UP000008820 UP000095301 UP000036403 UP000007819 UP000030765 UP000007801 UP000183832
UP000283053 UP000235965 UP000007266 UP000002358 UP000030742 UP000192223 UP000007798 UP000242457 UP000005203 UP000095300 UP000007755 UP000245037 UP000053105 UP000053825 UP000005205 UP000008792 UP000078492 UP000078540 UP000092462 UP000094527 UP000015103 UP000069940 UP000249989 UP000008820 UP000095301 UP000036403 UP000007819 UP000030765 UP000007801 UP000183832
Interpro
Gene 3D
ProteinModelPortal
H9JP83
A0A2H1WUP7
I4DP01
I4DLC0
A0A2A4JIX8
A0A0L7L6J1
+ More
A0A0N0PBN4 A0A212FHT2 A0A194QJM9 A0A0N1I833 I4DLB5 A0A194RFD5 A0A2A4JI81 H9JP17 A0A194QNP8 A0A194QNQ3 A0A212FHS6 A0A3S2LTM3 A0A2Z5U7W2 E0XEM8 A0A1Y1KUS1 A0A2J7PV34 D6WJC9 J3JZF0 A0A0L7L6D7 A0A2J7PV36 A0A0N1IES4 K7J5D9 A0A194QHX1 M9NHL6 A0A194QI87 U4TVX1 U5EWT8 A0A139WHG8 A0A194QHS8 A0A336KBK1 A0A1L8DLY9 A0A336KH96 A0A1B6DWB3 A0A1W4XKZ3 A0A1L8DM86 A0A0Q9X2K6 M9NK07 A0A1W6EW18 A0A2A3E259 A0A087ZSJ2 A0A1I8PKJ7 A0A194QJM0 A0A1W4XLV1 A0A0N1IP04 F4W8D2 A0A2P8Z9M7 V5I8P9 A0A0M8ZQU0 A0A0L7RA20 M4QVG7 M4QKU5 M4QII6 A0A158NKR2 B4MG80 A0A1B6G0U5 D6WJC8 Q9NDP1 A0A151IV06 A0A1B6GG13 A0A195BA50 A0A1B0DL94 A0A0Q9VYG4 A0A1D2NDD8 A0A1W6EWA9 M4QIK2 A0A1B6JPW2 R4G322 A0A1B6J451 M4QID0 T1HLA6 A0A182GDM5 Q9BID6 T1PDE1 Q16IH7 A0A1I8N509 A0A0J7KWI6 J9JXU7 A0A1B6LND9 A0A084VYL7 A0A1W4XBG2 A0A0C9PVI0 A0A1W4XBP4 A0A1Q3FSM1 D6WJC6 A0A2H8TR03 A0A0C9REN7 A0A1I8N523 A0A1W4XBF7 E9IDS8 J9LYB7 W8ANX0 T1HLF0 A0A0P8XYJ7 A0A1J1HLE1
A0A0N0PBN4 A0A212FHT2 A0A194QJM9 A0A0N1I833 I4DLB5 A0A194RFD5 A0A2A4JI81 H9JP17 A0A194QNP8 A0A194QNQ3 A0A212FHS6 A0A3S2LTM3 A0A2Z5U7W2 E0XEM8 A0A1Y1KUS1 A0A2J7PV34 D6WJC9 J3JZF0 A0A0L7L6D7 A0A2J7PV36 A0A0N1IES4 K7J5D9 A0A194QHX1 M9NHL6 A0A194QI87 U4TVX1 U5EWT8 A0A139WHG8 A0A194QHS8 A0A336KBK1 A0A1L8DLY9 A0A336KH96 A0A1B6DWB3 A0A1W4XKZ3 A0A1L8DM86 A0A0Q9X2K6 M9NK07 A0A1W6EW18 A0A2A3E259 A0A087ZSJ2 A0A1I8PKJ7 A0A194QJM0 A0A1W4XLV1 A0A0N1IP04 F4W8D2 A0A2P8Z9M7 V5I8P9 A0A0M8ZQU0 A0A0L7RA20 M4QVG7 M4QKU5 M4QII6 A0A158NKR2 B4MG80 A0A1B6G0U5 D6WJC8 Q9NDP1 A0A151IV06 A0A1B6GG13 A0A195BA50 A0A1B0DL94 A0A0Q9VYG4 A0A1D2NDD8 A0A1W6EWA9 M4QIK2 A0A1B6JPW2 R4G322 A0A1B6J451 M4QID0 T1HLA6 A0A182GDM5 Q9BID6 T1PDE1 Q16IH7 A0A1I8N509 A0A0J7KWI6 J9JXU7 A0A1B6LND9 A0A084VYL7 A0A1W4XBG2 A0A0C9PVI0 A0A1W4XBP4 A0A1Q3FSM1 D6WJC6 A0A2H8TR03 A0A0C9REN7 A0A1I8N523 A0A1W4XBF7 E9IDS8 J9LYB7 W8ANX0 T1HLF0 A0A0P8XYJ7 A0A1J1HLE1
PDB
5XFE
E-value=2.98562e-62,
Score=603
Ontologies
PATHWAY
GO
PANTHER
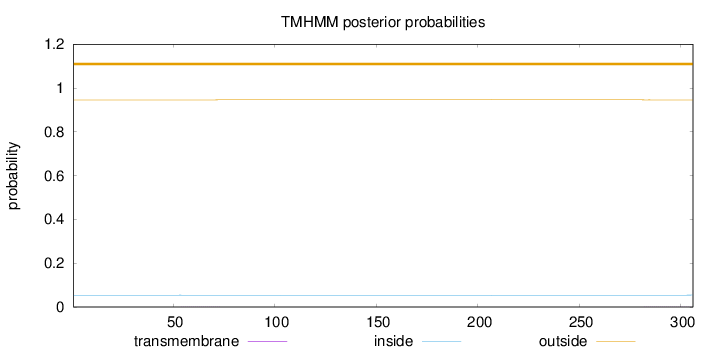
Topology
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02845
Exp number, first 60 AAs:
0.0017
Total prob of N-in:
0.05410
outside
1 - 306
Population Genetic Test Statistics
Pi
14.637545
Theta
22.527388
Tajima's D
-0.993321
CLR
51.015639
CSRT
0.141092945352732
Interpretation
Uncertain
