Gene
KWMTBOMO13969
Pre Gene Modal
BGIBMGA011278
Annotation
PREDICTED:_WD_and_tetratricopeptide_repeats_protein_1_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 1.379 Nuclear Reliability : 1.15
Sequence
CDS
ATGGGTAGGAGGGAGGAACAGGCATGGGCGAAGGGCACCTTAGGTCTGGCGGTGGCCCGAGAGCGAGGTGACATCGACAGGCAGTTCCAAAACCGATTAATGGTGACGAGGAGTATGATCGACAGACTTGGGTTGGAGAAAGAGCTTCAAGGGCACATGGGATGCGTCAACTGCCTAGAGTGGAATGCTGATGGATCAGTTCTCGCATCGGCATCAGATGATCTGCATGTTATTTTATGGGACCCGTACCGTTACAAGCAGCTACACAATATTGCTACGGGACACACTGGAAATATATTTTCAGTTAAATTTCTGTCGCCGGACACGATCGTGACGTGCGCGGCCGACAGCACGGTGCGGGCTCGCAGCGTGTGCGGGGGCGCCACGCTGCTGGAGTGCTCCTGCCACTGCGGCAGAGTCAAGAGGCTGGCCGCTCCTCCTGATACGCCTCATCTCATCTGGTCTGCGGGAGAAGACGGACTGATATTGCAGCACGACCTCCGCACGGCGCACCGCTGCAACTCTGACTCCTCGAATGTCCTGATTAACCTGCTGAACCACCTGGGCCGGTACGCCGAGGCCAAGTGCGTAGCCGTCAACCCCAGGAGGCCTTACCAGCTGGCCGTCGGTGCTAATGACTTTTACGTTCGCCTGTACGATACCAGGATGGTCAAGCTGGCCAAAATACAAACCAAGGCGTCGTCCACGTCGCACCACCGCATGTTGTGGGAGCGGCAGCACGTGTGCAGCTCGCGCGCCGGCGCCGGCGACCCGGAGAACAACATCCTCCGGGACTCCGTGCACTACTTCGCACCAGGTCACTTGTCCATGGAACCGAACGAGCACACCTTCCCCAAGAAGTCGACGACGTACGTGGCCTTCAGTAACGACGGAAACGAGTTGCTGGTGAACCTGGGCTCCGAGCAGGTGTACCTGTTCGACTTGAACAACGGTCGGAGCCCCGTCTTGCTGGAGAGCTTCATCATCCAACATAACCACGGCCCCAAGGACGGTAAGGTGGCTCCGAAGAATGGAACCAGCGTGCCCATCCTTGCCGACTCCCCTCCCCCCGCGCCCGCCGCGCCCGCGGCCCCCGTCGACTCGCCCACCCACCAGCTGCCAGACCATGTCAAGCAACTCAAGGAAACGGCGAACGGGTACATGGCAAAGGGGAACTACTCGCACGCCGTGGAGTACTACAACCGGGCGCTGGCGGAGTGCCAGAACTGCGCCATCCTGTACTCCAACCGAGCCGCCGCGCTCATGAGGAGGGGCTGGGCCGGGGACACGTACGCCGCCGTCCGGGACTGCTACACGGCCATCCGCCTCGAGCCGGCGCACGTCAAGTCGCACTTCCGGCTCGCGAAGGGGCTCATGGAGCTGCGCAAGGTGCGCGAGGCCCACGAGTCGCTGCTCTACTTCAAGGACAAGTTCCCGCGGCACGCCTCCAGCCACGCCGTGTTCCTGCTGCAGAAGGACATCAACACGCTCCTGGAGGCCAACGACGCGCAGCCCGCCGAAGGGTCCGGAAGTCCAGGCAGCGAAGAGGAGGGCACCGGGACGCAGATCTCGGCCTTGGAGAAGCAGCTCCGCGCCACGGCCCGCGACTACACGTGTCGGTTCCTGGGCCACTGCAACACCACCACGGACATCAAGGAGGCCAACTTCCTGGGGCCCGACGCCGAGTACATCGCCGCGGGCTCCGACGACGGCAGCATCTTCATCTGGAGTCGCCGCACCGGCAACATCGTGCGGTGTCTGCGCGGGGACGAGTCCATCGTCAACTGCATCCAGATGCACCCCAGCATGTTCCTGCTGGCCACCAGCGGCATCGAGGCCGTGGTGCGGCTGTGGAGCCCCCGCCCGGAGGTGAGTCCACCACGACGCTTTGCTGGTTTGTATTTACCAGAACAGTAA
Protein
MGRREEQAWAKGTLGLAVARERGDIDRQFQNRLMVTRSMIDRLGLEKELQGHMGCVNCLEWNADGSVLASASDDLHVILWDPYRYKQLHNIATGHTGNIFSVKFLSPDTIVTCAADSTVRARSVCGGATLLECSCHCGRVKRLAAPPDTPHLIWSAGEDGLILQHDLRTAHRCNSDSSNVLINLLNHLGRYAEAKCVAVNPRRPYQLAVGANDFYVRLYDTRMVKLAKIQTKASSTSHHRMLWERQHVCSSRAGAGDPENNILRDSVHYFAPGHLSMEPNEHTFPKKSTTYVAFSNDGNELLVNLGSEQVYLFDLNNGRSPVLLESFIIQHNHGPKDGKVAPKNGTSVPILADSPPPAPAAPAAPVDSPTHQLPDHVKQLKETANGYMAKGNYSHAVEYYNRALAECQNCAILYSNRAAALMRRGWAGDTYAAVRDCYTAIRLEPAHVKSHFRLAKGLMELRKVREAHESLLYFKDKFPRHASSHAVFLLQKDINTLLEANDAQPAEGSGSPGSEEEGTGTQISALEKQLRATARDYTCRFLGHCNTTTDIKEANFLGPDAEYIAAGSDDGSIFIWSRRTGNIVRCLRGDESIVNCIQMHPSMFLLATSGIEAVVRLWSPRPEVSPPRRFAGLYLPEQ
Summary
Uniprot
A0A2A4ITR6
A0A2A4ITM0
A0A2H1VR45
A0A3S2P2P9
F4WAW9
A0A026WZC3
+ More
A0A2A3E794 T1IVI1 A0A088A7K5 A0A0M8ZZS8 E2ABI0 A0A0L7R3D8 A0A1B6GAN1 E2C805 A0A147BTV8 A0A131Y1C1 A0A293N5W5 A0A1B6HEV9 A0A232EKI6 A0A1B6LG63 K7J1K8 A0A023FBT7 K1QJ92 A0A0L8H4Z2 A0A1B6D964 A0A0V0GAP1 A0A0K8TQT8 A0A1Q3G3H6 A0A1S4EUP3 A0A0A9XYU9 W5JDP3 C3YXJ5 A0A182NSR3 A0A182F7V2 A0A182TU18 A0A0K8UMF7 A0A182UYR0 A0A182JVP5 A0A182VY85 A7T282 A0A2I4D7T9 A0A2I4D7U4 A0A1S4GHP6 A0A1A8GKL8 A0A099YZC1 A0A182XKD6 A0A3Q2DDN6 A0A1A8KSS1 A0A1A8U1G7 A0A1A8DGA2 A0A3B5MBC3 A0A0S7KKR5 M4A8J8 A0A182HJK8 A0A146P4K6 A0A1A8B3V1 A0A182RDB0 A0A182R0J6 A0A3Q4I3K4 I3JI65 A0A3P9NUJ8 A0A1A8PT78 A0A1A8SKM2 K7F7L6 A0A3B3X054 A0A093I022 A0A3Q3CY86 A0A3B4F7P4 A0A1A7YU66 A0A087XWE5 A0A3B3W2L0 A0A3P8PPG8 A0A3P9CWR0 W5US09 A0A093NRI2 V9KJD6 A0A091T9S7 A0A1S3P6D8 A0A091RN91 A0A093D4H2 A0A091UAP1 R0LDW7 A0A091JGE2 A0A091NIC3 A0A091SM08 A0A087QW61 A0A093FY58 A0A091VJX7 A0A093EYG9 A0A087V231 A0A0A0AAP9 A0A091NXT1 U3J8L4 R7VSB3 A0A091FVJ7 W5M8M4
A0A2A3E794 T1IVI1 A0A088A7K5 A0A0M8ZZS8 E2ABI0 A0A0L7R3D8 A0A1B6GAN1 E2C805 A0A147BTV8 A0A131Y1C1 A0A293N5W5 A0A1B6HEV9 A0A232EKI6 A0A1B6LG63 K7J1K8 A0A023FBT7 K1QJ92 A0A0L8H4Z2 A0A1B6D964 A0A0V0GAP1 A0A0K8TQT8 A0A1Q3G3H6 A0A1S4EUP3 A0A0A9XYU9 W5JDP3 C3YXJ5 A0A182NSR3 A0A182F7V2 A0A182TU18 A0A0K8UMF7 A0A182UYR0 A0A182JVP5 A0A182VY85 A7T282 A0A2I4D7T9 A0A2I4D7U4 A0A1S4GHP6 A0A1A8GKL8 A0A099YZC1 A0A182XKD6 A0A3Q2DDN6 A0A1A8KSS1 A0A1A8U1G7 A0A1A8DGA2 A0A3B5MBC3 A0A0S7KKR5 M4A8J8 A0A182HJK8 A0A146P4K6 A0A1A8B3V1 A0A182RDB0 A0A182R0J6 A0A3Q4I3K4 I3JI65 A0A3P9NUJ8 A0A1A8PT78 A0A1A8SKM2 K7F7L6 A0A3B3X054 A0A093I022 A0A3Q3CY86 A0A3B4F7P4 A0A1A7YU66 A0A087XWE5 A0A3B3W2L0 A0A3P8PPG8 A0A3P9CWR0 W5US09 A0A093NRI2 V9KJD6 A0A091T9S7 A0A1S3P6D8 A0A091RN91 A0A093D4H2 A0A091UAP1 R0LDW7 A0A091JGE2 A0A091NIC3 A0A091SM08 A0A087QW61 A0A093FY58 A0A091VJX7 A0A093EYG9 A0A087V231 A0A0A0AAP9 A0A091NXT1 U3J8L4 R7VSB3 A0A091FVJ7 W5M8M4
Pubmed
EMBL
NWSH01007141
PCG63069.1
PCG63071.1
ODYU01003774
SOQ42952.1
RSAL01000042
+ More
RVE50822.1 GL888054 EGI68659.1 KK107078 QOIP01000011 EZA60494.1 RLU16828.1 KZ288347 PBC27570.1 JH431584 KQ435789 KOX74450.1 GL438255 EFN69207.1 KQ414663 KOC65400.1 GECZ01010289 JAS59480.1 GL453506 EFN75923.1 GEGO01001216 JAR94188.1 GEFM01002737 JAP73059.1 GFWV01023072 MAA47799.1 GECU01034500 JAS73206.1 NNAY01003809 OXU18828.1 GEBQ01017358 JAT22619.1 GBBI01000309 JAC18403.1 JH817051 EKC21711.1 KQ419180 KOF84366.1 GEDC01015163 GEDC01008492 GEDC01007206 JAS22135.1 JAS28806.1 JAS30092.1 GECL01001003 JAP05121.1 GDAI01001090 JAI16513.1 GFDL01000688 JAV34357.1 GBHO01021134 GDHC01001904 JAG22470.1 JAQ16725.1 ADMH02001520 ETN62216.1 GG666563 EEN54804.1 GDHF01024420 GDHF01002855 JAI27894.1 JAI49459.1 DS470212 EDO29936.1 AAAB01008904 HAEB01013382 HAEC01003466 SBQ71543.1 KL887292 KGL74796.1 HAEE01015196 SBR35246.1 HAEJ01001687 SBS42144.1 HADZ01017818 HAEA01003429 SBQ31909.1 GBYX01206115 GBYX01206114 JAO77893.1 APCN01002179 GCES01147570 GCES01146554 JAQ38752.1 HADY01022670 SBP61155.1 AXCN02000227 AERX01027683 HAEF01015081 HAEG01009317 SBR84199.1 HAEI01015979 SBS18448.1 AGCU01160105 AGCU01160106 AGCU01160107 KL206708 KFV85160.1 HADW01008792 HADX01011501 SBP33733.1 AYCK01007860 JT417498 AHH42398.1 KL224693 KFW62747.1 JW865702 AFO98219.1 KK443771 KFQ71036.1 KK705418 KFQ29977.1 KL474107 KFV19979.1 KK425898 KFQ87729.1 KB743278 EOA99719.1 KK500853 KFP10801.1 KL383792 KFP89196.1 KK478239 KFQ59679.1 KL225955 KFM05465.1 KK399883 KFV59331.1 KL411121 KFR03439.1 KK612528 KFV46944.1 KL477013 KFO06673.1 KL871162 KGL90972.1 KK643935 KFP97662.1 ADON01086184 KB375351 AKCR02000266 EMC78694.1 PKK17822.1 KL447465 KFO73673.1 AHAT01009549 AHAT01009550
RVE50822.1 GL888054 EGI68659.1 KK107078 QOIP01000011 EZA60494.1 RLU16828.1 KZ288347 PBC27570.1 JH431584 KQ435789 KOX74450.1 GL438255 EFN69207.1 KQ414663 KOC65400.1 GECZ01010289 JAS59480.1 GL453506 EFN75923.1 GEGO01001216 JAR94188.1 GEFM01002737 JAP73059.1 GFWV01023072 MAA47799.1 GECU01034500 JAS73206.1 NNAY01003809 OXU18828.1 GEBQ01017358 JAT22619.1 GBBI01000309 JAC18403.1 JH817051 EKC21711.1 KQ419180 KOF84366.1 GEDC01015163 GEDC01008492 GEDC01007206 JAS22135.1 JAS28806.1 JAS30092.1 GECL01001003 JAP05121.1 GDAI01001090 JAI16513.1 GFDL01000688 JAV34357.1 GBHO01021134 GDHC01001904 JAG22470.1 JAQ16725.1 ADMH02001520 ETN62216.1 GG666563 EEN54804.1 GDHF01024420 GDHF01002855 JAI27894.1 JAI49459.1 DS470212 EDO29936.1 AAAB01008904 HAEB01013382 HAEC01003466 SBQ71543.1 KL887292 KGL74796.1 HAEE01015196 SBR35246.1 HAEJ01001687 SBS42144.1 HADZ01017818 HAEA01003429 SBQ31909.1 GBYX01206115 GBYX01206114 JAO77893.1 APCN01002179 GCES01147570 GCES01146554 JAQ38752.1 HADY01022670 SBP61155.1 AXCN02000227 AERX01027683 HAEF01015081 HAEG01009317 SBR84199.1 HAEI01015979 SBS18448.1 AGCU01160105 AGCU01160106 AGCU01160107 KL206708 KFV85160.1 HADW01008792 HADX01011501 SBP33733.1 AYCK01007860 JT417498 AHH42398.1 KL224693 KFW62747.1 JW865702 AFO98219.1 KK443771 KFQ71036.1 KK705418 KFQ29977.1 KL474107 KFV19979.1 KK425898 KFQ87729.1 KB743278 EOA99719.1 KK500853 KFP10801.1 KL383792 KFP89196.1 KK478239 KFQ59679.1 KL225955 KFM05465.1 KK399883 KFV59331.1 KL411121 KFR03439.1 KK612528 KFV46944.1 KL477013 KFO06673.1 KL871162 KGL90972.1 KK643935 KFP97662.1 ADON01086184 KB375351 AKCR02000266 EMC78694.1 PKK17822.1 KL447465 KFO73673.1 AHAT01009549 AHAT01009550
Proteomes
UP000218220
UP000283053
UP000007755
UP000053097
UP000279307
UP000242457
+ More
UP000005203 UP000053105 UP000000311 UP000053825 UP000008237 UP000215335 UP000002358 UP000005408 UP000053454 UP000000673 UP000001554 UP000075884 UP000069272 UP000075902 UP000075903 UP000075881 UP000075920 UP000001593 UP000192220 UP000053641 UP000076407 UP000265020 UP000261380 UP000002852 UP000075840 UP000265000 UP000075900 UP000075886 UP000261580 UP000005207 UP000242638 UP000007267 UP000261480 UP000053584 UP000264840 UP000261460 UP000028760 UP000261500 UP000265100 UP000265160 UP000221080 UP000054081 UP000087266 UP000053119 UP000053286 UP000053283 UP000053858 UP000016666 UP000053872 UP000053760 UP000018468
UP000005203 UP000053105 UP000000311 UP000053825 UP000008237 UP000215335 UP000002358 UP000005408 UP000053454 UP000000673 UP000001554 UP000075884 UP000069272 UP000075902 UP000075903 UP000075881 UP000075920 UP000001593 UP000192220 UP000053641 UP000076407 UP000265020 UP000261380 UP000002852 UP000075840 UP000265000 UP000075900 UP000075886 UP000261580 UP000005207 UP000242638 UP000007267 UP000261480 UP000053584 UP000264840 UP000261460 UP000028760 UP000261500 UP000265100 UP000265160 UP000221080 UP000054081 UP000087266 UP000053119 UP000053286 UP000053283 UP000053858 UP000016666 UP000053872 UP000053760 UP000018468
Interpro
Gene 3D
ProteinModelPortal
A0A2A4ITR6
A0A2A4ITM0
A0A2H1VR45
A0A3S2P2P9
F4WAW9
A0A026WZC3
+ More
A0A2A3E794 T1IVI1 A0A088A7K5 A0A0M8ZZS8 E2ABI0 A0A0L7R3D8 A0A1B6GAN1 E2C805 A0A147BTV8 A0A131Y1C1 A0A293N5W5 A0A1B6HEV9 A0A232EKI6 A0A1B6LG63 K7J1K8 A0A023FBT7 K1QJ92 A0A0L8H4Z2 A0A1B6D964 A0A0V0GAP1 A0A0K8TQT8 A0A1Q3G3H6 A0A1S4EUP3 A0A0A9XYU9 W5JDP3 C3YXJ5 A0A182NSR3 A0A182F7V2 A0A182TU18 A0A0K8UMF7 A0A182UYR0 A0A182JVP5 A0A182VY85 A7T282 A0A2I4D7T9 A0A2I4D7U4 A0A1S4GHP6 A0A1A8GKL8 A0A099YZC1 A0A182XKD6 A0A3Q2DDN6 A0A1A8KSS1 A0A1A8U1G7 A0A1A8DGA2 A0A3B5MBC3 A0A0S7KKR5 M4A8J8 A0A182HJK8 A0A146P4K6 A0A1A8B3V1 A0A182RDB0 A0A182R0J6 A0A3Q4I3K4 I3JI65 A0A3P9NUJ8 A0A1A8PT78 A0A1A8SKM2 K7F7L6 A0A3B3X054 A0A093I022 A0A3Q3CY86 A0A3B4F7P4 A0A1A7YU66 A0A087XWE5 A0A3B3W2L0 A0A3P8PPG8 A0A3P9CWR0 W5US09 A0A093NRI2 V9KJD6 A0A091T9S7 A0A1S3P6D8 A0A091RN91 A0A093D4H2 A0A091UAP1 R0LDW7 A0A091JGE2 A0A091NIC3 A0A091SM08 A0A087QW61 A0A093FY58 A0A091VJX7 A0A093EYG9 A0A087V231 A0A0A0AAP9 A0A091NXT1 U3J8L4 R7VSB3 A0A091FVJ7 W5M8M4
A0A2A3E794 T1IVI1 A0A088A7K5 A0A0M8ZZS8 E2ABI0 A0A0L7R3D8 A0A1B6GAN1 E2C805 A0A147BTV8 A0A131Y1C1 A0A293N5W5 A0A1B6HEV9 A0A232EKI6 A0A1B6LG63 K7J1K8 A0A023FBT7 K1QJ92 A0A0L8H4Z2 A0A1B6D964 A0A0V0GAP1 A0A0K8TQT8 A0A1Q3G3H6 A0A1S4EUP3 A0A0A9XYU9 W5JDP3 C3YXJ5 A0A182NSR3 A0A182F7V2 A0A182TU18 A0A0K8UMF7 A0A182UYR0 A0A182JVP5 A0A182VY85 A7T282 A0A2I4D7T9 A0A2I4D7U4 A0A1S4GHP6 A0A1A8GKL8 A0A099YZC1 A0A182XKD6 A0A3Q2DDN6 A0A1A8KSS1 A0A1A8U1G7 A0A1A8DGA2 A0A3B5MBC3 A0A0S7KKR5 M4A8J8 A0A182HJK8 A0A146P4K6 A0A1A8B3V1 A0A182RDB0 A0A182R0J6 A0A3Q4I3K4 I3JI65 A0A3P9NUJ8 A0A1A8PT78 A0A1A8SKM2 K7F7L6 A0A3B3X054 A0A093I022 A0A3Q3CY86 A0A3B4F7P4 A0A1A7YU66 A0A087XWE5 A0A3B3W2L0 A0A3P8PPG8 A0A3P9CWR0 W5US09 A0A093NRI2 V9KJD6 A0A091T9S7 A0A1S3P6D8 A0A091RN91 A0A093D4H2 A0A091UAP1 R0LDW7 A0A091JGE2 A0A091NIC3 A0A091SM08 A0A087QW61 A0A093FY58 A0A091VJX7 A0A093EYG9 A0A087V231 A0A0A0AAP9 A0A091NXT1 U3J8L4 R7VSB3 A0A091FVJ7 W5M8M4
PDB
4GCO
E-value=5.60031e-08,
Score=139
Ontologies
GO
Topology
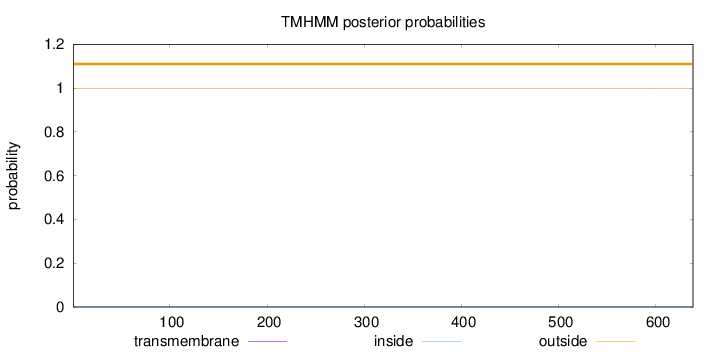
Length:
638
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02326
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00116
outside
1 - 638
Population Genetic Test Statistics
Pi
284.660235
Theta
185.533031
Tajima's D
1.650886
CLR
0.295748
CSRT
0.819909004549773
Interpretation
Uncertain
