Pre Gene Modal
BGIBMGA011281
Annotation
trafficking_protein_particle_complex_subunit_5_[Papilio_xuthus]
Full name
Trafficking protein particle complex subunit 5
Location in the cell
Mitochondrial Reliability : 2.036
Sequence
CDS
ATGAACATGTCCTCCAGTCGTTCGAAATCATCAATATTAGATAAACCATTACGAAAAGGGAAAGGGGAGGTCTCTTTAGCCTTTTATGCCCTTTTATTTTCTGAAATTGTTCAGTATAGTCAGAATCGATCGCATTCGATACAAGATCTACAGTTTAAATTGTCTGAAATCGGACAAGATGTGGGCACGAGACTGCTCGACTTGTATTTTGTGAGAGAGAGAAATAGCAAACGTGAGATAAAACTGCTGAATATGTTACTGTTTATCAAGTCTACTTTATGGAAGGTGCTTTTTGGTAAAGAAGCTGATAAACTGGAACATGCAAATGATGATGAAAGGACCTACTATATTATCGAGAAAGATGCTCTTGTAAATAAGTTTATAAGTGTACCAAAAGACAAGGGTTCATTAAACTGTGCCATCTTCAATGCTGGCATCATTGAAGCTGTTTTAACTAGGAGCGGATTTCCAGCAAAGGTGACAGCACACTGGCACAAGGGAACAACATACATGGTCAAGTTCGATGATTCAGTAATCACCAGAGACAAAACATTGGATGATCGTTAA
Protein
MNMSSSRSKSSILDKPLRKGKGEVSLAFYALLFSEIVQYSQNRSHSIQDLQFKLSEIGQDVGTRLLDLYFVRERNSKREIKLLNMLLFIKSTLWKVLFGKEADKLEHANDDERTYYIIEKDALVNKFISVPKDKGSLNCAIFNAGIIEAVLTRSGFPAKVTAHWHKGTTYMVKFDDSVITRDKTLDDR
Summary
Description
May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Subunit
Part of the multisubunit TRAPP (transport protein particle) complex.
Similarity
Belongs to the TRAPP small subunits family. BET3 subfamily.
Uniprot
H9JP27
I4DLB7
A0A2A4K3E2
A0A2H1W9Z2
L7X1T5
S4NX29
+ More
C3PPG5 A0A212FD17 A0A0K8TNL6 A0A1L8DVY4 A0A067R3H0 A0A2J7PV46 A0A1B0CQT9 A0A1Y1L632 A0A182RSV3 A0A182WD17 A0A182YCA7 A0A0C9RRV8 A0A232F1Q5 K7JAW6 A0A224XLC0 A0A0V0G8Z6 A0A1B6CK81 A0A182N3P7 D7ELH8 W5JII0 A0A0P4VRR0 R4FKT2 A0A182P1D9 A0A182V5Z7 A0A182TTV3 A0A182X080 A0A182KTB6 Q7PDE6 A0A182HPJ6 A0A182JUN8 A0A023EIG4 A0A310SDT0 A0A2M3ZD55 A0A2M4AXT1 A0A182FTW4 A0A2M4C0E5 A0A182QKP5 A0A195DT05 A0A151JT55 E2AMW5 F4W7F0 A0A195ATC4 A0A158NPU4 A0A151X8N1 A0A1B6M110 A0A1Q3FGG7 A0A0J7KXI0 U5EY29 E2BQG5 Q17H04 A0A1B6H8B6 A0A1B6FDZ1 B0WI08 A0A3L8DA56 A0A026X218 A0A182IVG2 A0A088ARP0 A0A2A3E3M6 A0A1A9W8E5 C3Z0P3 A0A0M8ZWQ8 B3ME28 A0A0M4E5Y4 A0A1B0G7V2 A0A1A9VQ68 Q291B6 B4GAY3 A0A1W4UHK8 A0A0J9RE19 B4P7N8 B4HRP2 B3NQT0 Q7K2Q8 A0A154PA58 A0A2R7Y1Y8 A0A2H8TZ62 A0A0L7RD14 C4WVR1 B4MNS7 B4LM74 B4KNN6 A0A2S2PRP6 A0A3B0J696 A0A034VQD3 A0A0A9YTL2 A0A151I6Y8 B4J6K8 Q5TMY9 A0A1I8MAE4 A0A0A1XGY7 A0A1I8P538 E0VWU3 R7UIT4 A0A1S3JI87
C3PPG5 A0A212FD17 A0A0K8TNL6 A0A1L8DVY4 A0A067R3H0 A0A2J7PV46 A0A1B0CQT9 A0A1Y1L632 A0A182RSV3 A0A182WD17 A0A182YCA7 A0A0C9RRV8 A0A232F1Q5 K7JAW6 A0A224XLC0 A0A0V0G8Z6 A0A1B6CK81 A0A182N3P7 D7ELH8 W5JII0 A0A0P4VRR0 R4FKT2 A0A182P1D9 A0A182V5Z7 A0A182TTV3 A0A182X080 A0A182KTB6 Q7PDE6 A0A182HPJ6 A0A182JUN8 A0A023EIG4 A0A310SDT0 A0A2M3ZD55 A0A2M4AXT1 A0A182FTW4 A0A2M4C0E5 A0A182QKP5 A0A195DT05 A0A151JT55 E2AMW5 F4W7F0 A0A195ATC4 A0A158NPU4 A0A151X8N1 A0A1B6M110 A0A1Q3FGG7 A0A0J7KXI0 U5EY29 E2BQG5 Q17H04 A0A1B6H8B6 A0A1B6FDZ1 B0WI08 A0A3L8DA56 A0A026X218 A0A182IVG2 A0A088ARP0 A0A2A3E3M6 A0A1A9W8E5 C3Z0P3 A0A0M8ZWQ8 B3ME28 A0A0M4E5Y4 A0A1B0G7V2 A0A1A9VQ68 Q291B6 B4GAY3 A0A1W4UHK8 A0A0J9RE19 B4P7N8 B4HRP2 B3NQT0 Q7K2Q8 A0A154PA58 A0A2R7Y1Y8 A0A2H8TZ62 A0A0L7RD14 C4WVR1 B4MNS7 B4LM74 B4KNN6 A0A2S2PRP6 A0A3B0J696 A0A034VQD3 A0A0A9YTL2 A0A151I6Y8 B4J6K8 Q5TMY9 A0A1I8MAE4 A0A0A1XGY7 A0A1I8P538 E0VWU3 R7UIT4 A0A1S3JI87
Pubmed
19121390
22651552
26354079
23674305
23622113
22118469
+ More
26369729 24845553 28004739 25244985 28648823 20075255 18362917 19820115 20920257 23761445 27129103 20966253 12364791 14747013 17210077 24945155 20798317 21719571 21347285 17510324 30249741 24508170 18563158 17994087 15632085 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25401762 26823975 25315136 25830018 20566863 23254933
26369729 24845553 28004739 25244985 28648823 20075255 18362917 19820115 20920257 23761445 27129103 20966253 12364791 14747013 17210077 24945155 20798317 21719571 21347285 17510324 30249741 24508170 18563158 17994087 15632085 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25401762 26823975 25315136 25830018 20566863 23254933
EMBL
BABH01009604
AK402085
KQ458761
BAM18707.1
KPJ05137.1
NWSH01000204
+ More
PCG78438.1 ODYU01007143 SOQ49662.1 KC469894 AGC92743.1 GAIX01009239 JAA83321.1 CU681835 CU672275 CAY54173.1 AGBW02009117 OWR51634.1 GDAI01001644 JAI15959.1 GFDF01003629 JAV10455.1 KK853019 KDR12406.1 NEVH01020989 PNF20201.1 AJWK01024033 GEZM01063659 JAV69034.1 GBYB01011280 JAG81047.1 NNAY01001286 OXU24482.1 AAZX01011201 GFTR01003140 JAW13286.1 GECL01001497 JAP04627.1 GEDC01023409 JAS13889.1 DS497866 EFA12180.1 ADMH02001378 ETN62705.1 GDKW01001473 JAI55122.1 ACPB03017117 GAHY01001713 JAA75797.1 AAAB01008986 EAA00628.5 APCN01003261 GAPW01004701 JAC08897.1 KQ760869 OAD58759.1 GGFM01005639 MBW26390.1 GGFK01012259 MBW45580.1 GGFJ01009645 MBW58786.1 AXCN02001102 KQ980487 KYN15922.1 KQ982014 KYN30636.1 GL440936 EFN65239.1 GL887844 EGI69823.1 KQ976745 KYM75473.1 ADTU01022754 KQ982409 KYQ56722.1 GEBQ01010372 JAT29605.1 GFDL01008398 JAV26647.1 LBMM01002308 KMQ95003.1 GANO01002225 JAB57646.1 GL449769 EFN82023.1 CH477254 EAT45920.1 GECU01036853 JAS70853.1 GECZ01021323 JAS48446.1 DS231941 EDS28090.1 QOIP01000011 RLU17003.1 KK107021 EZA62332.1 KZ288427 PBC25856.1 GG666569 EEN53923.1 KQ435814 KOX72665.1 CH902619 EDV37573.1 CP012524 ALC41791.1 CCAG010021503 CM000071 EAL25196.1 CH479181 EDW32085.1 CM002911 KMY93834.1 CM000158 EDW91064.1 CH480816 EDW47906.1 CH954179 EDV55990.1 AE013599 AY060730 AAF58209.1 AAL28278.1 KQ434856 KZC08723.1 KK854080 PUA16085.1 GFXV01007798 MBW19603.1 KQ414614 KOC68852.1 ABLF02013830 AK341629 BAH71981.1 CH963848 EDW73766.1 CH940648 EDW60952.1 CH933808 EDW10021.1 GGMR01019523 MBY32142.1 OUUW01000001 SPP75292.1 GAKP01014283 JAC44669.1 GBHO01009181 GBRD01000935 GDHC01001519 JAG34423.1 JAG64886.1 JAQ17110.1 KQ978456 KYM93894.1 CH916367 EDW00911.1 EAA00657.4 GBXI01004102 JAD10190.1 DS235824 EEB17849.1 AMQN01008362 KB302959 ELU03713.1
PCG78438.1 ODYU01007143 SOQ49662.1 KC469894 AGC92743.1 GAIX01009239 JAA83321.1 CU681835 CU672275 CAY54173.1 AGBW02009117 OWR51634.1 GDAI01001644 JAI15959.1 GFDF01003629 JAV10455.1 KK853019 KDR12406.1 NEVH01020989 PNF20201.1 AJWK01024033 GEZM01063659 JAV69034.1 GBYB01011280 JAG81047.1 NNAY01001286 OXU24482.1 AAZX01011201 GFTR01003140 JAW13286.1 GECL01001497 JAP04627.1 GEDC01023409 JAS13889.1 DS497866 EFA12180.1 ADMH02001378 ETN62705.1 GDKW01001473 JAI55122.1 ACPB03017117 GAHY01001713 JAA75797.1 AAAB01008986 EAA00628.5 APCN01003261 GAPW01004701 JAC08897.1 KQ760869 OAD58759.1 GGFM01005639 MBW26390.1 GGFK01012259 MBW45580.1 GGFJ01009645 MBW58786.1 AXCN02001102 KQ980487 KYN15922.1 KQ982014 KYN30636.1 GL440936 EFN65239.1 GL887844 EGI69823.1 KQ976745 KYM75473.1 ADTU01022754 KQ982409 KYQ56722.1 GEBQ01010372 JAT29605.1 GFDL01008398 JAV26647.1 LBMM01002308 KMQ95003.1 GANO01002225 JAB57646.1 GL449769 EFN82023.1 CH477254 EAT45920.1 GECU01036853 JAS70853.1 GECZ01021323 JAS48446.1 DS231941 EDS28090.1 QOIP01000011 RLU17003.1 KK107021 EZA62332.1 KZ288427 PBC25856.1 GG666569 EEN53923.1 KQ435814 KOX72665.1 CH902619 EDV37573.1 CP012524 ALC41791.1 CCAG010021503 CM000071 EAL25196.1 CH479181 EDW32085.1 CM002911 KMY93834.1 CM000158 EDW91064.1 CH480816 EDW47906.1 CH954179 EDV55990.1 AE013599 AY060730 AAF58209.1 AAL28278.1 KQ434856 KZC08723.1 KK854080 PUA16085.1 GFXV01007798 MBW19603.1 KQ414614 KOC68852.1 ABLF02013830 AK341629 BAH71981.1 CH963848 EDW73766.1 CH940648 EDW60952.1 CH933808 EDW10021.1 GGMR01019523 MBY32142.1 OUUW01000001 SPP75292.1 GAKP01014283 JAC44669.1 GBHO01009181 GBRD01000935 GDHC01001519 JAG34423.1 JAG64886.1 JAQ17110.1 KQ978456 KYM93894.1 CH916367 EDW00911.1 EAA00657.4 GBXI01004102 JAD10190.1 DS235824 EEB17849.1 AMQN01008362 KB302959 ELU03713.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000027135
UP000235965
+ More
UP000092461 UP000075900 UP000075920 UP000076408 UP000215335 UP000002358 UP000075884 UP000007266 UP000000673 UP000015103 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000069272 UP000075886 UP000078492 UP000078541 UP000000311 UP000007755 UP000078540 UP000005205 UP000075809 UP000036403 UP000008237 UP000008820 UP000002320 UP000279307 UP000053097 UP000075880 UP000005203 UP000242457 UP000091820 UP000001554 UP000053105 UP000007801 UP000092553 UP000092444 UP000078200 UP000001819 UP000008744 UP000192221 UP000002282 UP000001292 UP000008711 UP000000803 UP000076502 UP000053825 UP000007819 UP000007798 UP000008792 UP000009192 UP000268350 UP000078542 UP000001070 UP000095301 UP000095300 UP000009046 UP000014760 UP000085678
UP000092461 UP000075900 UP000075920 UP000076408 UP000215335 UP000002358 UP000075884 UP000007266 UP000000673 UP000015103 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000069272 UP000075886 UP000078492 UP000078541 UP000000311 UP000007755 UP000078540 UP000005205 UP000075809 UP000036403 UP000008237 UP000008820 UP000002320 UP000279307 UP000053097 UP000075880 UP000005203 UP000242457 UP000091820 UP000001554 UP000053105 UP000007801 UP000092553 UP000092444 UP000078200 UP000001819 UP000008744 UP000192221 UP000002282 UP000001292 UP000008711 UP000000803 UP000076502 UP000053825 UP000007819 UP000007798 UP000008792 UP000009192 UP000268350 UP000078542 UP000001070 UP000095301 UP000095300 UP000009046 UP000014760 UP000085678
Pfam
PF04051 TRAPP
Interpro
SUPFAM
SSF111126
SSF111126
ProteinModelPortal
H9JP27
I4DLB7
A0A2A4K3E2
A0A2H1W9Z2
L7X1T5
S4NX29
+ More
C3PPG5 A0A212FD17 A0A0K8TNL6 A0A1L8DVY4 A0A067R3H0 A0A2J7PV46 A0A1B0CQT9 A0A1Y1L632 A0A182RSV3 A0A182WD17 A0A182YCA7 A0A0C9RRV8 A0A232F1Q5 K7JAW6 A0A224XLC0 A0A0V0G8Z6 A0A1B6CK81 A0A182N3P7 D7ELH8 W5JII0 A0A0P4VRR0 R4FKT2 A0A182P1D9 A0A182V5Z7 A0A182TTV3 A0A182X080 A0A182KTB6 Q7PDE6 A0A182HPJ6 A0A182JUN8 A0A023EIG4 A0A310SDT0 A0A2M3ZD55 A0A2M4AXT1 A0A182FTW4 A0A2M4C0E5 A0A182QKP5 A0A195DT05 A0A151JT55 E2AMW5 F4W7F0 A0A195ATC4 A0A158NPU4 A0A151X8N1 A0A1B6M110 A0A1Q3FGG7 A0A0J7KXI0 U5EY29 E2BQG5 Q17H04 A0A1B6H8B6 A0A1B6FDZ1 B0WI08 A0A3L8DA56 A0A026X218 A0A182IVG2 A0A088ARP0 A0A2A3E3M6 A0A1A9W8E5 C3Z0P3 A0A0M8ZWQ8 B3ME28 A0A0M4E5Y4 A0A1B0G7V2 A0A1A9VQ68 Q291B6 B4GAY3 A0A1W4UHK8 A0A0J9RE19 B4P7N8 B4HRP2 B3NQT0 Q7K2Q8 A0A154PA58 A0A2R7Y1Y8 A0A2H8TZ62 A0A0L7RD14 C4WVR1 B4MNS7 B4LM74 B4KNN6 A0A2S2PRP6 A0A3B0J696 A0A034VQD3 A0A0A9YTL2 A0A151I6Y8 B4J6K8 Q5TMY9 A0A1I8MAE4 A0A0A1XGY7 A0A1I8P538 E0VWU3 R7UIT4 A0A1S3JI87
C3PPG5 A0A212FD17 A0A0K8TNL6 A0A1L8DVY4 A0A067R3H0 A0A2J7PV46 A0A1B0CQT9 A0A1Y1L632 A0A182RSV3 A0A182WD17 A0A182YCA7 A0A0C9RRV8 A0A232F1Q5 K7JAW6 A0A224XLC0 A0A0V0G8Z6 A0A1B6CK81 A0A182N3P7 D7ELH8 W5JII0 A0A0P4VRR0 R4FKT2 A0A182P1D9 A0A182V5Z7 A0A182TTV3 A0A182X080 A0A182KTB6 Q7PDE6 A0A182HPJ6 A0A182JUN8 A0A023EIG4 A0A310SDT0 A0A2M3ZD55 A0A2M4AXT1 A0A182FTW4 A0A2M4C0E5 A0A182QKP5 A0A195DT05 A0A151JT55 E2AMW5 F4W7F0 A0A195ATC4 A0A158NPU4 A0A151X8N1 A0A1B6M110 A0A1Q3FGG7 A0A0J7KXI0 U5EY29 E2BQG5 Q17H04 A0A1B6H8B6 A0A1B6FDZ1 B0WI08 A0A3L8DA56 A0A026X218 A0A182IVG2 A0A088ARP0 A0A2A3E3M6 A0A1A9W8E5 C3Z0P3 A0A0M8ZWQ8 B3ME28 A0A0M4E5Y4 A0A1B0G7V2 A0A1A9VQ68 Q291B6 B4GAY3 A0A1W4UHK8 A0A0J9RE19 B4P7N8 B4HRP2 B3NQT0 Q7K2Q8 A0A154PA58 A0A2R7Y1Y8 A0A2H8TZ62 A0A0L7RD14 C4WVR1 B4MNS7 B4LM74 B4KNN6 A0A2S2PRP6 A0A3B0J696 A0A034VQD3 A0A0A9YTL2 A0A151I6Y8 B4J6K8 Q5TMY9 A0A1I8MAE4 A0A0A1XGY7 A0A1I8P538 E0VWU3 R7UIT4 A0A1S3JI87
PDB
2J3W
E-value=1.31733e-63,
Score=612
Ontologies
GO
PANTHER
Topology
Subcellular location
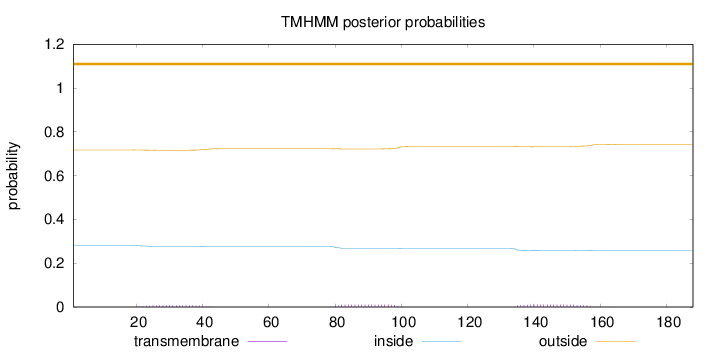
Length:
188
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.57588
Exp number, first 60 AAs:
0.14352
Total prob of N-in:
0.28243
outside
1 - 188
Population Genetic Test Statistics
Pi
190.540465
Theta
184.607013
Tajima's D
-0.719679
CLR
1172.401902
CSRT
0.193240337983101
Interpretation
Uncertain
