Pre Gene Modal
BGIBMGA011323
Annotation
PREDICTED:_uncharacterized_protein_LOC101746156_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.859
Sequence
CDS
ATGGCAAATAACTACGGCTTATCTGATGCAGAACTCAACCTTATTAAAACCCAAGCCTCAAGGCGTGCTGAAATGAGACGGGAGTTCTTGAAACAAAGAACAAACCCCTGGAAAAACGCTTCGGAGGCTGGTTATGTTTTCGACACAGCATTGCAAAGGTTTCTGTCCATGAAAGTAACACAGTTTGAATATTTTACCGTAAACAAGCGAACTAGTTTGTTTGGATTTTTTGTTATTGTTGTACCCATGTTTACATTTGGTACCTTGATATGGAACGAGAGAACGCAGCGTGAACAGAAGATTCGCAGTGGAGAGCTCCGTTACAAAGACAGACTCTTCAAGCTAGCTTAA
Protein
MANNYGLSDAELNLIKTQASRRAEMRREFLKQRTNPWKNASEAGYVFDTALQRFLSMKVTQFEYFTVNKRTSLFGFFVIVVPMFTFGTLIWNERTQREQKIRSGELRYKDRLFKLA
Summary
Uniprot
H9JP69
A0A0L7KQ87
S4NN57
A0A1E1WEQ9
A0A3S2NY67
A0A212FD21
+ More
A0A2A4K2G7 C3PPG4 A0A194RFV4 A0A194QHV6 L7WWD5 A0A0L7KQB4 A0A2H1W9G6 A0A336MY24 A0A3S2TJ86 R4V2S5 V5GYY5 A0A2T7NUU2 A0A212FD38 A0A067QTX1 A0A1W4X571 C4WU79 A0A3R7PYG9 A0A2S2QV90 A0A2H8TKX4 A0A1A9W8E7 A0A1A9VQ69 A0A1W4U6I9 N6U0U6 Q7PZB8 A0A182UYK2 A0A182ULF0 A0A182XGS5 A0A182HHE2 A0A2M4C491 W5JPI2 A0A182JDS0 A0A084WIU2 A0A1B0G7V3 T1E7W7 A0A2M3ZGX9 A0A1A9XF04 A0A1B0ARD3 B4MNS6 A0A182N3W4 A0A182L4X0 A0A0V0G600 A0A182P7I0 A0A182FUZ9 R4WQ72 A0A170W1S1 A0A069DN95 Q6IDF5 B4HRP1 A0A182W1X5 A0A2M4AAZ9 A0A0J9RCQ5 Q17H03 A0A182QAP8 A0A1B0ADC0 A0A1D2MBU3 B3NQT1 Q71DD3 A0A2M4AB98 A0A182LRE9 A0A1B0CQU0 A0A1J1IHU4 A0A0P4VZI1 R4G4D9 B3ME29 W8BVS7 A0A182RYY2 A0A182YLA0 A0A023F7K3 A0A1I8NC06 A0A3B0JRI0 A0A182JXK3 U5ERR7 A0A1I8PEC8 Q291B7 B4GAY2 A0A1L8EEY5 E9H0B0 A0A023ED18 A0A182TCS3 A0A2R5LFB1 A0A0K8VTL2 A0A0M4EKA4 A0A224XQM6 A0A131XNP1 A0A1L8EF10 A0A1L8D9K7 A0A034VT01 B4KNN5 B4LM75 A0A023EF07 A0A2R7X8F0 A0A023EF00 A0A0L0BRH9 A0A1L8E8U0
A0A2A4K2G7 C3PPG4 A0A194RFV4 A0A194QHV6 L7WWD5 A0A0L7KQB4 A0A2H1W9G6 A0A336MY24 A0A3S2TJ86 R4V2S5 V5GYY5 A0A2T7NUU2 A0A212FD38 A0A067QTX1 A0A1W4X571 C4WU79 A0A3R7PYG9 A0A2S2QV90 A0A2H8TKX4 A0A1A9W8E7 A0A1A9VQ69 A0A1W4U6I9 N6U0U6 Q7PZB8 A0A182UYK2 A0A182ULF0 A0A182XGS5 A0A182HHE2 A0A2M4C491 W5JPI2 A0A182JDS0 A0A084WIU2 A0A1B0G7V3 T1E7W7 A0A2M3ZGX9 A0A1A9XF04 A0A1B0ARD3 B4MNS6 A0A182N3W4 A0A182L4X0 A0A0V0G600 A0A182P7I0 A0A182FUZ9 R4WQ72 A0A170W1S1 A0A069DN95 Q6IDF5 B4HRP1 A0A182W1X5 A0A2M4AAZ9 A0A0J9RCQ5 Q17H03 A0A182QAP8 A0A1B0ADC0 A0A1D2MBU3 B3NQT1 Q71DD3 A0A2M4AB98 A0A182LRE9 A0A1B0CQU0 A0A1J1IHU4 A0A0P4VZI1 R4G4D9 B3ME29 W8BVS7 A0A182RYY2 A0A182YLA0 A0A023F7K3 A0A1I8NC06 A0A3B0JRI0 A0A182JXK3 U5ERR7 A0A1I8PEC8 Q291B7 B4GAY2 A0A1L8EEY5 E9H0B0 A0A023ED18 A0A182TCS3 A0A2R5LFB1 A0A0K8VTL2 A0A0M4EKA4 A0A224XQM6 A0A131XNP1 A0A1L8EF10 A0A1L8D9K7 A0A034VT01 B4KNN5 B4LM75 A0A023EF07 A0A2R7X8F0 A0A023EF00 A0A0L0BRH9 A0A1L8E8U0
Pubmed
19121390
26227816
23622113
22118469
26354079
23674305
+ More
24845553 23537049 12364791 14747013 17210077 20920257 23761445 24438588 17994087 20966253 23691247 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17510324 27289101 14525923 17550304 27129103 24495485 25244985 25474469 25315136 15632085 21292972 24945155 28049606 25348373 26483478 26108605
24845553 23537049 12364791 14747013 17210077 20920257 23761445 24438588 17994087 20966253 23691247 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17510324 27289101 14525923 17550304 27129103 24495485 25244985 25474469 25315136 15632085 21292972 24945155 28049606 25348373 26483478 26108605
EMBL
BABH01009600
JTDY01007372
KOB65260.1
GAIX01012369
JAA80191.1
GDQN01005615
+ More
JAT85439.1 RSAL01000099 RVE47567.1 AGBW02009117 OWR51632.1 NWSH01000204 PCG78437.1 CU681835 CU672275 CAY54172.1 KQ460297 KPJ16185.1 KQ458761 KPJ05138.1 KC469894 AGC92742.1 KOB65261.1 ODYU01007143 SOQ49663.1 UFQS01000092 UFQS01001485 UFQT01000092 UFQT01001485 SSW99194.1 SSX30758.1 RVE47568.1 KC632249 AGM32063.1 GALX01001369 JAB67097.1 PZQS01000009 PVD24906.1 OWR51633.1 KK853019 KDR12405.1 ABLF02016923 AK340947 BAH71449.1 QCYY01001042 ROT80985.1 GGMS01012474 MBY81677.1 GFXV01002133 MBW13938.1 APGK01053959 KB741239 KB632275 ENN72167.1 ERL91136.1 AAAB01008986 EAA00737.4 APCN01002407 GGFJ01010973 MBW60114.1 ADMH02000915 ETN64674.1 ATLV01023946 KE525347 KFB50136.1 CCAG010021503 GAMD01003288 JAA98302.1 GGFM01007065 MBW27816.1 JXJN01002362 CH963848 EDW73765.1 GECL01002976 JAP03148.1 AK417841 BAN21056.1 GEMB01006210 JAR97122.1 GBGD01003594 JAC85295.1 AE013599 BT014650 AAF58210.1 AAT27274.1 CH480816 EDW47905.1 GGFK01004634 MBW37955.1 CM002911 KMY93833.1 CH477254 EAT45921.1 AXCN02001095 LJIJ01001948 ODM90468.1 CH954179 EDV55991.1 AF531936 CM000158 AAQ09835.1 EDW91063.1 GGFK01004743 MBW38064.1 AXCM01000489 AJWK01024033 CVRI01000053 CRK99831.1 GDKW01001652 JAI54943.1 ACPB03002356 GAHY01001450 JAA76060.1 CH902619 EDV37574.1 GAMC01003233 JAC03323.1 GBBI01001494 JAC17218.1 OUUW01000001 SPP75291.1 GANO01003599 JAB56272.1 CM000071 EAL25195.1 CH479181 EDW32084.1 GFDG01001575 JAV17224.1 GL732580 EFX74848.1 GAPW01006285 JAC07313.1 GGLE01004033 MBY08159.1 GDHF01010098 GDHF01006500 GDHF01002541 JAI42216.1 JAI45814.1 JAI49773.1 CP012524 ALC41792.1 GFTR01001619 JAW14807.1 GEFH01000629 JAP67952.1 GFDG01001574 JAV17225.1 GFDF01010935 JAV03149.1 GAKP01014284 JAC44668.1 CH933808 EDW10020.1 CH940648 EDW60953.1 JXUM01081565 GAPW01006284 KQ563255 JAC07314.1 KXJ74193.1 KK857445 PTY27175.1 GAPW01006283 JAC07315.1 JRES01001473 KNC22626.1 GFDG01003621 JAV15178.1
JAT85439.1 RSAL01000099 RVE47567.1 AGBW02009117 OWR51632.1 NWSH01000204 PCG78437.1 CU681835 CU672275 CAY54172.1 KQ460297 KPJ16185.1 KQ458761 KPJ05138.1 KC469894 AGC92742.1 KOB65261.1 ODYU01007143 SOQ49663.1 UFQS01000092 UFQS01001485 UFQT01000092 UFQT01001485 SSW99194.1 SSX30758.1 RVE47568.1 KC632249 AGM32063.1 GALX01001369 JAB67097.1 PZQS01000009 PVD24906.1 OWR51633.1 KK853019 KDR12405.1 ABLF02016923 AK340947 BAH71449.1 QCYY01001042 ROT80985.1 GGMS01012474 MBY81677.1 GFXV01002133 MBW13938.1 APGK01053959 KB741239 KB632275 ENN72167.1 ERL91136.1 AAAB01008986 EAA00737.4 APCN01002407 GGFJ01010973 MBW60114.1 ADMH02000915 ETN64674.1 ATLV01023946 KE525347 KFB50136.1 CCAG010021503 GAMD01003288 JAA98302.1 GGFM01007065 MBW27816.1 JXJN01002362 CH963848 EDW73765.1 GECL01002976 JAP03148.1 AK417841 BAN21056.1 GEMB01006210 JAR97122.1 GBGD01003594 JAC85295.1 AE013599 BT014650 AAF58210.1 AAT27274.1 CH480816 EDW47905.1 GGFK01004634 MBW37955.1 CM002911 KMY93833.1 CH477254 EAT45921.1 AXCN02001095 LJIJ01001948 ODM90468.1 CH954179 EDV55991.1 AF531936 CM000158 AAQ09835.1 EDW91063.1 GGFK01004743 MBW38064.1 AXCM01000489 AJWK01024033 CVRI01000053 CRK99831.1 GDKW01001652 JAI54943.1 ACPB03002356 GAHY01001450 JAA76060.1 CH902619 EDV37574.1 GAMC01003233 JAC03323.1 GBBI01001494 JAC17218.1 OUUW01000001 SPP75291.1 GANO01003599 JAB56272.1 CM000071 EAL25195.1 CH479181 EDW32084.1 GFDG01001575 JAV17224.1 GL732580 EFX74848.1 GAPW01006285 JAC07313.1 GGLE01004033 MBY08159.1 GDHF01010098 GDHF01006500 GDHF01002541 JAI42216.1 JAI45814.1 JAI49773.1 CP012524 ALC41792.1 GFTR01001619 JAW14807.1 GEFH01000629 JAP67952.1 GFDG01001574 JAV17225.1 GFDF01010935 JAV03149.1 GAKP01014284 JAC44668.1 CH933808 EDW10020.1 CH940648 EDW60953.1 JXUM01081565 GAPW01006284 KQ563255 JAC07314.1 KXJ74193.1 KK857445 PTY27175.1 GAPW01006283 JAC07315.1 JRES01001473 KNC22626.1 GFDG01003621 JAV15178.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000007151
UP000218220
UP000053240
+ More
UP000053268 UP000245119 UP000027135 UP000192223 UP000007819 UP000283509 UP000091820 UP000078200 UP000192221 UP000019118 UP000030742 UP000007062 UP000075903 UP000075902 UP000076407 UP000075840 UP000000673 UP000075880 UP000030765 UP000092444 UP000092443 UP000092460 UP000007798 UP000075884 UP000075882 UP000075885 UP000069272 UP000000803 UP000001292 UP000075920 UP000008820 UP000075886 UP000092445 UP000094527 UP000008711 UP000002282 UP000075883 UP000092461 UP000183832 UP000015103 UP000007801 UP000075900 UP000076408 UP000095301 UP000268350 UP000075881 UP000095300 UP000001819 UP000008744 UP000000305 UP000075901 UP000092553 UP000009192 UP000008792 UP000069940 UP000249989 UP000037069
UP000053268 UP000245119 UP000027135 UP000192223 UP000007819 UP000283509 UP000091820 UP000078200 UP000192221 UP000019118 UP000030742 UP000007062 UP000075903 UP000075902 UP000076407 UP000075840 UP000000673 UP000075880 UP000030765 UP000092444 UP000092443 UP000092460 UP000007798 UP000075884 UP000075882 UP000075885 UP000069272 UP000000803 UP000001292 UP000075920 UP000008820 UP000075886 UP000092445 UP000094527 UP000008711 UP000002282 UP000075883 UP000092461 UP000183832 UP000015103 UP000007801 UP000075900 UP000076408 UP000095301 UP000268350 UP000075881 UP000095300 UP000001819 UP000008744 UP000000305 UP000075901 UP000092553 UP000009192 UP000008792 UP000069940 UP000249989 UP000037069
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JP69
A0A0L7KQ87
S4NN57
A0A1E1WEQ9
A0A3S2NY67
A0A212FD21
+ More
A0A2A4K2G7 C3PPG4 A0A194RFV4 A0A194QHV6 L7WWD5 A0A0L7KQB4 A0A2H1W9G6 A0A336MY24 A0A3S2TJ86 R4V2S5 V5GYY5 A0A2T7NUU2 A0A212FD38 A0A067QTX1 A0A1W4X571 C4WU79 A0A3R7PYG9 A0A2S2QV90 A0A2H8TKX4 A0A1A9W8E7 A0A1A9VQ69 A0A1W4U6I9 N6U0U6 Q7PZB8 A0A182UYK2 A0A182ULF0 A0A182XGS5 A0A182HHE2 A0A2M4C491 W5JPI2 A0A182JDS0 A0A084WIU2 A0A1B0G7V3 T1E7W7 A0A2M3ZGX9 A0A1A9XF04 A0A1B0ARD3 B4MNS6 A0A182N3W4 A0A182L4X0 A0A0V0G600 A0A182P7I0 A0A182FUZ9 R4WQ72 A0A170W1S1 A0A069DN95 Q6IDF5 B4HRP1 A0A182W1X5 A0A2M4AAZ9 A0A0J9RCQ5 Q17H03 A0A182QAP8 A0A1B0ADC0 A0A1D2MBU3 B3NQT1 Q71DD3 A0A2M4AB98 A0A182LRE9 A0A1B0CQU0 A0A1J1IHU4 A0A0P4VZI1 R4G4D9 B3ME29 W8BVS7 A0A182RYY2 A0A182YLA0 A0A023F7K3 A0A1I8NC06 A0A3B0JRI0 A0A182JXK3 U5ERR7 A0A1I8PEC8 Q291B7 B4GAY2 A0A1L8EEY5 E9H0B0 A0A023ED18 A0A182TCS3 A0A2R5LFB1 A0A0K8VTL2 A0A0M4EKA4 A0A224XQM6 A0A131XNP1 A0A1L8EF10 A0A1L8D9K7 A0A034VT01 B4KNN5 B4LM75 A0A023EF07 A0A2R7X8F0 A0A023EF00 A0A0L0BRH9 A0A1L8E8U0
A0A2A4K2G7 C3PPG4 A0A194RFV4 A0A194QHV6 L7WWD5 A0A0L7KQB4 A0A2H1W9G6 A0A336MY24 A0A3S2TJ86 R4V2S5 V5GYY5 A0A2T7NUU2 A0A212FD38 A0A067QTX1 A0A1W4X571 C4WU79 A0A3R7PYG9 A0A2S2QV90 A0A2H8TKX4 A0A1A9W8E7 A0A1A9VQ69 A0A1W4U6I9 N6U0U6 Q7PZB8 A0A182UYK2 A0A182ULF0 A0A182XGS5 A0A182HHE2 A0A2M4C491 W5JPI2 A0A182JDS0 A0A084WIU2 A0A1B0G7V3 T1E7W7 A0A2M3ZGX9 A0A1A9XF04 A0A1B0ARD3 B4MNS6 A0A182N3W4 A0A182L4X0 A0A0V0G600 A0A182P7I0 A0A182FUZ9 R4WQ72 A0A170W1S1 A0A069DN95 Q6IDF5 B4HRP1 A0A182W1X5 A0A2M4AAZ9 A0A0J9RCQ5 Q17H03 A0A182QAP8 A0A1B0ADC0 A0A1D2MBU3 B3NQT1 Q71DD3 A0A2M4AB98 A0A182LRE9 A0A1B0CQU0 A0A1J1IHU4 A0A0P4VZI1 R4G4D9 B3ME29 W8BVS7 A0A182RYY2 A0A182YLA0 A0A023F7K3 A0A1I8NC06 A0A3B0JRI0 A0A182JXK3 U5ERR7 A0A1I8PEC8 Q291B7 B4GAY2 A0A1L8EEY5 E9H0B0 A0A023ED18 A0A182TCS3 A0A2R5LFB1 A0A0K8VTL2 A0A0M4EKA4 A0A224XQM6 A0A131XNP1 A0A1L8EF10 A0A1L8D9K7 A0A034VT01 B4KNN5 B4LM75 A0A023EF07 A0A2R7X8F0 A0A023EF00 A0A0L0BRH9 A0A1L8E8U0
PDB
5LNK
E-value=0.0154666,
Score=83
Ontologies
PATHWAY
GO
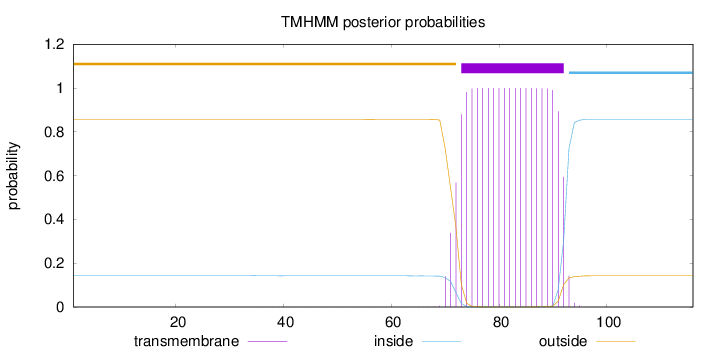
Topology
Length:
116
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.57407
Exp number, first 60 AAs:
0.02239
Total prob of N-in:
0.14326
outside
1 - 72
TMhelix
73 - 92
inside
93 - 116
Population Genetic Test Statistics
Pi
100.892166
Theta
156.595584
Tajima's D
-1.131071
CLR
0
CSRT
0.11754412279386
Interpretation
Uncertain
