Gene
KWMTBOMO13959
Pre Gene Modal
BGIBMGA011322
Annotation
unnamed_protein_product_[Heliconius_melpomene]
Full name
TM2 domain-containing protein CG11103
Location in the cell
PlasmaMembrane Reliability : 2.318
Sequence
CDS
ATGGCACTGATAACAAAGTCAACAGCTGTAACAGTATTGTTTATAACAGGGATAATTAAGGCAAAATCTGAAAGCGATCCAATTTCAAAAGAACCATATAGGCCACTCGGTCCTTTAGTTAAATGTTCATATCTACCGCTGGAATTCCTAGATTGCGACGAACCTATCGACCATAAAGGGAATCACACCGCCCGGCAAGCCACAGGCCATGGTTGCGTCAAATTTGGAGGTGTCCGATATGATCAAGTTGAGAAAGCTAAAGTTCAGTGTAAAGCCTTGGATGGAATTGAATGTTATGGCAGTCGAAGCTTTCCGCGTGACGGTTTCCCGTGTGTGCGATACTCTGGACATTACTTTACGACCACCTTGATTTATAGCATCTTGTTAGGATTCCTAGGTATGGATAGATTTTGTCTTGGTCAAACAGGAACAGCTGTGGGGAAACTTTTGACCCTAGGAGGGCTTGGTATTTGGTGGATCGTTGATGTAGTGTTATTGATTACCAACAGCCTACACCCTGAAGATGGTAGTAATTGGAATCCATATGTTTAA
Protein
MALITKSTAVTVLFITGIIKAKSESDPISKEPYRPLGPLVKCSYLPLEFLDCDEPIDHKGNHTARQATGHGCVKFGGVRYDQVEKAKVQCKALDGIECYGSRSFPRDGFPCVRYSGHYFTTTLIYSILLGFLGMDRFCLGQTGTAVGKLLTLGGLGIWWIVDVVLLITNSLHPEDGSNWNPYV
Summary
Similarity
Belongs to the TM2 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain TM2 domain-containing protein CG11103
Uniprot
H9JP68
A0A2A4K2G6
A0A0L7KQM0
A0A212FD29
A0A194QNU3
C3PPH9
+ More
L7X405 A0A194RFG3 C3PPG2 A0A3S2L7N9 S4P0I2 A0A1E1WKH5 A0A182RNX0 A0A182LXG0 A0A182GKL0 B0WFA8 Q16RN1 A0A1Q3FAH3 A0A1B6MFS9 A0A182TC14 A0A182Y453 A0A182W007 A0A182K8N5 A0A182V7T9 A0A182TKY8 A0A182LGI3 A0A182XHQ0 Q7QCV7 A0A182HTN7 A0A1B6D840 A0A224XYJ8 N6T211 T1I1S6 A0A0J7L5B3 A0A088ATQ1 A0A182P267 A0A232EY23 A0A3L8DWL8 A0A151XAE3 A0A026X4R5 K7J9A2 A0A195B7R4 A0A158NGQ2 F4WZY2 E1ZV61 A0A1B6HC11 A0A195FFR8 E9ILC1 A0A084W7C9 A0A0M8ZYS4 A0A1B6GYM7 A0A310SRU0 A0A195DXN9 D6WCG6 B4L387 E2BDA1 A0A2A3E5S4 E0VN94 A0A0A9WT83 A0A0P5HBT0 A0A182JFN0 A0A1Y1KEP6 A0A1W4XF55 A0A0P5FDX7 A0A2H1WQU3 A0A0L7QLP3 W5J159 E9GLT9 W8CB25 A0A0A1WGR6 A0A0K8WIW8 A0A182NKT4 A0A0K8WJN5 A0A1Y9G9R4 B4M3R4 A0A1V9WZA0 A0A151IFF8 A0A2M4C0M3 A0A1W4VSV2 B4JXL7 A0A034WPH4 A7SBH8 A0A2M4AZC7 A0A0M4ESN8 Q29HU7 B4GY88 B4Q2G3 A0A3B0JWK1 Q9VY86 B4R4K0 B4IG70 B3NVY8 B4NCJ1 A0A1S3I3P1 B3N121 A0A2J7QSG7 A0A0L8ICV5 A0A0K8RGU1 A0A293M527 A0A0C9S4Z9
L7X405 A0A194RFG3 C3PPG2 A0A3S2L7N9 S4P0I2 A0A1E1WKH5 A0A182RNX0 A0A182LXG0 A0A182GKL0 B0WFA8 Q16RN1 A0A1Q3FAH3 A0A1B6MFS9 A0A182TC14 A0A182Y453 A0A182W007 A0A182K8N5 A0A182V7T9 A0A182TKY8 A0A182LGI3 A0A182XHQ0 Q7QCV7 A0A182HTN7 A0A1B6D840 A0A224XYJ8 N6T211 T1I1S6 A0A0J7L5B3 A0A088ATQ1 A0A182P267 A0A232EY23 A0A3L8DWL8 A0A151XAE3 A0A026X4R5 K7J9A2 A0A195B7R4 A0A158NGQ2 F4WZY2 E1ZV61 A0A1B6HC11 A0A195FFR8 E9ILC1 A0A084W7C9 A0A0M8ZYS4 A0A1B6GYM7 A0A310SRU0 A0A195DXN9 D6WCG6 B4L387 E2BDA1 A0A2A3E5S4 E0VN94 A0A0A9WT83 A0A0P5HBT0 A0A182JFN0 A0A1Y1KEP6 A0A1W4XF55 A0A0P5FDX7 A0A2H1WQU3 A0A0L7QLP3 W5J159 E9GLT9 W8CB25 A0A0A1WGR6 A0A0K8WIW8 A0A182NKT4 A0A0K8WJN5 A0A1Y9G9R4 B4M3R4 A0A1V9WZA0 A0A151IFF8 A0A2M4C0M3 A0A1W4VSV2 B4JXL7 A0A034WPH4 A7SBH8 A0A2M4AZC7 A0A0M4ESN8 Q29HU7 B4GY88 B4Q2G3 A0A3B0JWK1 Q9VY86 B4R4K0 B4IG70 B3NVY8 B4NCJ1 A0A1S3I3P1 B3N121 A0A2J7QSG7 A0A0L8ICV5 A0A0K8RGU1 A0A293M527 A0A0C9S4Z9
Pubmed
19121390
26227816
22118469
26354079
23674305
23622113
+ More
26483478 17510324 25244985 20966253 12364791 14747013 17210077 23537049 28648823 30249741 24508170 20075255 21347285 21719571 20798317 21282665 24438588 18362917 19820115 17994087 20566863 25401762 26823975 28004739 20920257 23761445 21292972 24495485 25830018 28327890 25348373 17615350 15632085 17550304 10731132 12537572 12537569 26131772
26483478 17510324 25244985 20966253 12364791 14747013 17210077 23537049 28648823 30249741 24508170 20075255 21347285 21719571 20798317 21282665 24438588 18362917 19820115 17994087 20566863 25401762 26823975 28004739 20920257 23761445 21292972 24495485 25830018 28327890 25348373 17615350 15632085 17550304 10731132 12537572 12537569 26131772
EMBL
BABH01009600
NWSH01000204
PCG78451.1
JTDY01007372
KOB65259.1
AGBW02009117
+ More
OWR51630.1 KQ458761 KPJ05151.1 CU681835 CAY54152.1 KC469894 AGC92740.1 KQ460297 KPJ16199.1 CU672275 CAY54170.1 RSAL01000099 RVE47565.1 GAIX01012930 JAA79630.1 GDQN01003550 JAT87504.1 AXCM01004114 JXUM01070018 JXUM01070019 JXUM01103694 KQ564826 KXJ71693.1 DS231916 EDS26125.1 CH477701 EAT37083.1 GFDL01010469 JAV24576.1 GEBQ01005222 JAT34755.1 AAAB01008859 EAA07714.5 APCN01000570 GEDC01015435 JAS21863.1 GFTR01003133 JAW13293.1 APGK01055379 KB741264 KB632006 ENN71548.1 ERL87981.1 ACPB03016273 LBMM01000692 KMQ97738.1 NNAY01001707 OXU23170.1 QOIP01000004 RLU24178.1 KQ982339 KYQ57332.1 KK107020 EZA62414.1 KQ976574 KYM80287.1 ADTU01015169 GL888480 EGI60272.1 GL434441 EFN74920.1 GECU01035537 JAS72169.1 KQ981625 KYN39072.1 GL764074 EFZ18641.1 ATLV01021211 KE525314 KFB46123.1 KQ435812 KOX72836.1 GECZ01002243 JAS67526.1 KQ760559 OAD59802.1 KQ980107 KYN17638.1 KQ971311 EEZ98875.1 CH933810 EDW07015.2 GL447585 EFN86329.1 KZ288357 PBC27113.1 DS235335 EEB14850.1 GBHO01033321 GBHO01032592 GBHO01032591 GBHO01032589 GBHO01032588 GBHO01032585 GBHO01013332 GBHO01010033 GBHO01010032 GBHO01010031 GBRD01010693 GBRD01010692 GBRD01010691 GDHC01018429 GDHC01010759 JAG10283.1 JAG11012.1 JAG11013.1 JAG11015.1 JAG11016.1 JAG11019.1 JAG30272.1 JAG33571.1 JAG33572.1 JAG33573.1 JAG55131.1 JAQ00200.1 JAQ07870.1 GDIP01144457 GDIQ01259453 GDIQ01230118 GDIQ01202526 GDIQ01134803 LRGB01000944 JAJ78945.1 JAK21607.1 KZS14490.1 AXCP01007741 GEZM01085671 JAV59952.1 GDIQ01256934 JAJ94790.1 ODYU01010352 SOQ55398.1 KQ414915 KOC59469.1 ADMH02002195 ETN57877.1 GL732551 EFX79627.1 GAMC01000839 JAC05717.1 GBXI01016083 JAC98208.1 GDHF01001277 JAI51037.1 GDHF01012178 GDHF01000953 JAI40136.1 JAI51361.1 CH940651 EDW65439.1 MNPL01032057 OQR66537.1 KQ977793 KYM99649.1 GGFJ01009719 MBW58860.1 CH916376 EDV95116.1 GAKP01002760 JAC56192.1 DS469616 EDO38925.1 GGFK01012809 MBW46130.1 CP012528 ALC49365.1 CH379064 EAL31660.2 CH479197 EDW27744.1 CM000162 EDX01624.1 OUUW01000003 SPP78099.1 AE014298 AY119007 CM000366 EDX17854.1 CH480835 EDW48804.1 CH954180 EDV47153.1 CH964239 EDW82550.1 CH902650 EDV30056.2 NEVH01011876 PNF31513.1 KQ415990 KOF99282.1 GADI01003512 JAA70296.1 GFWV01010462 MAA35191.1 GBZX01000326 JAG92414.1
OWR51630.1 KQ458761 KPJ05151.1 CU681835 CAY54152.1 KC469894 AGC92740.1 KQ460297 KPJ16199.1 CU672275 CAY54170.1 RSAL01000099 RVE47565.1 GAIX01012930 JAA79630.1 GDQN01003550 JAT87504.1 AXCM01004114 JXUM01070018 JXUM01070019 JXUM01103694 KQ564826 KXJ71693.1 DS231916 EDS26125.1 CH477701 EAT37083.1 GFDL01010469 JAV24576.1 GEBQ01005222 JAT34755.1 AAAB01008859 EAA07714.5 APCN01000570 GEDC01015435 JAS21863.1 GFTR01003133 JAW13293.1 APGK01055379 KB741264 KB632006 ENN71548.1 ERL87981.1 ACPB03016273 LBMM01000692 KMQ97738.1 NNAY01001707 OXU23170.1 QOIP01000004 RLU24178.1 KQ982339 KYQ57332.1 KK107020 EZA62414.1 KQ976574 KYM80287.1 ADTU01015169 GL888480 EGI60272.1 GL434441 EFN74920.1 GECU01035537 JAS72169.1 KQ981625 KYN39072.1 GL764074 EFZ18641.1 ATLV01021211 KE525314 KFB46123.1 KQ435812 KOX72836.1 GECZ01002243 JAS67526.1 KQ760559 OAD59802.1 KQ980107 KYN17638.1 KQ971311 EEZ98875.1 CH933810 EDW07015.2 GL447585 EFN86329.1 KZ288357 PBC27113.1 DS235335 EEB14850.1 GBHO01033321 GBHO01032592 GBHO01032591 GBHO01032589 GBHO01032588 GBHO01032585 GBHO01013332 GBHO01010033 GBHO01010032 GBHO01010031 GBRD01010693 GBRD01010692 GBRD01010691 GDHC01018429 GDHC01010759 JAG10283.1 JAG11012.1 JAG11013.1 JAG11015.1 JAG11016.1 JAG11019.1 JAG30272.1 JAG33571.1 JAG33572.1 JAG33573.1 JAG55131.1 JAQ00200.1 JAQ07870.1 GDIP01144457 GDIQ01259453 GDIQ01230118 GDIQ01202526 GDIQ01134803 LRGB01000944 JAJ78945.1 JAK21607.1 KZS14490.1 AXCP01007741 GEZM01085671 JAV59952.1 GDIQ01256934 JAJ94790.1 ODYU01010352 SOQ55398.1 KQ414915 KOC59469.1 ADMH02002195 ETN57877.1 GL732551 EFX79627.1 GAMC01000839 JAC05717.1 GBXI01016083 JAC98208.1 GDHF01001277 JAI51037.1 GDHF01012178 GDHF01000953 JAI40136.1 JAI51361.1 CH940651 EDW65439.1 MNPL01032057 OQR66537.1 KQ977793 KYM99649.1 GGFJ01009719 MBW58860.1 CH916376 EDV95116.1 GAKP01002760 JAC56192.1 DS469616 EDO38925.1 GGFK01012809 MBW46130.1 CP012528 ALC49365.1 CH379064 EAL31660.2 CH479197 EDW27744.1 CM000162 EDX01624.1 OUUW01000003 SPP78099.1 AE014298 AY119007 CM000366 EDX17854.1 CH480835 EDW48804.1 CH954180 EDV47153.1 CH964239 EDW82550.1 CH902650 EDV30056.2 NEVH01011876 PNF31513.1 KQ415990 KOF99282.1 GADI01003512 JAA70296.1 GFWV01010462 MAA35191.1 GBZX01000326 JAG92414.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000283053 UP000075900 UP000075883 UP000069940 UP000249989 UP000002320 UP000008820 UP000075901 UP000076408 UP000075920 UP000075881 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000019118 UP000030742 UP000015103 UP000036403 UP000005203 UP000075885 UP000215335 UP000279307 UP000075809 UP000053097 UP000002358 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000030765 UP000053105 UP000078492 UP000007266 UP000009192 UP000008237 UP000242457 UP000009046 UP000076858 UP000075880 UP000192223 UP000053825 UP000000673 UP000000305 UP000075884 UP000069272 UP000008792 UP000192247 UP000078542 UP000192221 UP000001070 UP000001593 UP000092553 UP000001819 UP000008744 UP000002282 UP000268350 UP000000803 UP000000304 UP000001292 UP000008711 UP000007798 UP000085678 UP000007801 UP000235965 UP000053454
UP000283053 UP000075900 UP000075883 UP000069940 UP000249989 UP000002320 UP000008820 UP000075901 UP000076408 UP000075920 UP000075881 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000019118 UP000030742 UP000015103 UP000036403 UP000005203 UP000075885 UP000215335 UP000279307 UP000075809 UP000053097 UP000002358 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000030765 UP000053105 UP000078492 UP000007266 UP000009192 UP000008237 UP000242457 UP000009046 UP000076858 UP000075880 UP000192223 UP000053825 UP000000673 UP000000305 UP000075884 UP000069272 UP000008792 UP000192247 UP000078542 UP000192221 UP000001070 UP000001593 UP000092553 UP000001819 UP000008744 UP000002282 UP000268350 UP000000803 UP000000304 UP000001292 UP000008711 UP000007798 UP000085678 UP000007801 UP000235965 UP000053454
PRIDE
ProteinModelPortal
H9JP68
A0A2A4K2G6
A0A0L7KQM0
A0A212FD29
A0A194QNU3
C3PPH9
+ More
L7X405 A0A194RFG3 C3PPG2 A0A3S2L7N9 S4P0I2 A0A1E1WKH5 A0A182RNX0 A0A182LXG0 A0A182GKL0 B0WFA8 Q16RN1 A0A1Q3FAH3 A0A1B6MFS9 A0A182TC14 A0A182Y453 A0A182W007 A0A182K8N5 A0A182V7T9 A0A182TKY8 A0A182LGI3 A0A182XHQ0 Q7QCV7 A0A182HTN7 A0A1B6D840 A0A224XYJ8 N6T211 T1I1S6 A0A0J7L5B3 A0A088ATQ1 A0A182P267 A0A232EY23 A0A3L8DWL8 A0A151XAE3 A0A026X4R5 K7J9A2 A0A195B7R4 A0A158NGQ2 F4WZY2 E1ZV61 A0A1B6HC11 A0A195FFR8 E9ILC1 A0A084W7C9 A0A0M8ZYS4 A0A1B6GYM7 A0A310SRU0 A0A195DXN9 D6WCG6 B4L387 E2BDA1 A0A2A3E5S4 E0VN94 A0A0A9WT83 A0A0P5HBT0 A0A182JFN0 A0A1Y1KEP6 A0A1W4XF55 A0A0P5FDX7 A0A2H1WQU3 A0A0L7QLP3 W5J159 E9GLT9 W8CB25 A0A0A1WGR6 A0A0K8WIW8 A0A182NKT4 A0A0K8WJN5 A0A1Y9G9R4 B4M3R4 A0A1V9WZA0 A0A151IFF8 A0A2M4C0M3 A0A1W4VSV2 B4JXL7 A0A034WPH4 A7SBH8 A0A2M4AZC7 A0A0M4ESN8 Q29HU7 B4GY88 B4Q2G3 A0A3B0JWK1 Q9VY86 B4R4K0 B4IG70 B3NVY8 B4NCJ1 A0A1S3I3P1 B3N121 A0A2J7QSG7 A0A0L8ICV5 A0A0K8RGU1 A0A293M527 A0A0C9S4Z9
L7X405 A0A194RFG3 C3PPG2 A0A3S2L7N9 S4P0I2 A0A1E1WKH5 A0A182RNX0 A0A182LXG0 A0A182GKL0 B0WFA8 Q16RN1 A0A1Q3FAH3 A0A1B6MFS9 A0A182TC14 A0A182Y453 A0A182W007 A0A182K8N5 A0A182V7T9 A0A182TKY8 A0A182LGI3 A0A182XHQ0 Q7QCV7 A0A182HTN7 A0A1B6D840 A0A224XYJ8 N6T211 T1I1S6 A0A0J7L5B3 A0A088ATQ1 A0A182P267 A0A232EY23 A0A3L8DWL8 A0A151XAE3 A0A026X4R5 K7J9A2 A0A195B7R4 A0A158NGQ2 F4WZY2 E1ZV61 A0A1B6HC11 A0A195FFR8 E9ILC1 A0A084W7C9 A0A0M8ZYS4 A0A1B6GYM7 A0A310SRU0 A0A195DXN9 D6WCG6 B4L387 E2BDA1 A0A2A3E5S4 E0VN94 A0A0A9WT83 A0A0P5HBT0 A0A182JFN0 A0A1Y1KEP6 A0A1W4XF55 A0A0P5FDX7 A0A2H1WQU3 A0A0L7QLP3 W5J159 E9GLT9 W8CB25 A0A0A1WGR6 A0A0K8WIW8 A0A182NKT4 A0A0K8WJN5 A0A1Y9G9R4 B4M3R4 A0A1V9WZA0 A0A151IFF8 A0A2M4C0M3 A0A1W4VSV2 B4JXL7 A0A034WPH4 A7SBH8 A0A2M4AZC7 A0A0M4ESN8 Q29HU7 B4GY88 B4Q2G3 A0A3B0JWK1 Q9VY86 B4R4K0 B4IG70 B3NVY8 B4NCJ1 A0A1S3I3P1 B3N121 A0A2J7QSG7 A0A0L8ICV5 A0A0K8RGU1 A0A293M527 A0A0C9S4Z9
Ontologies
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 21,
Likelihood: 0.715033
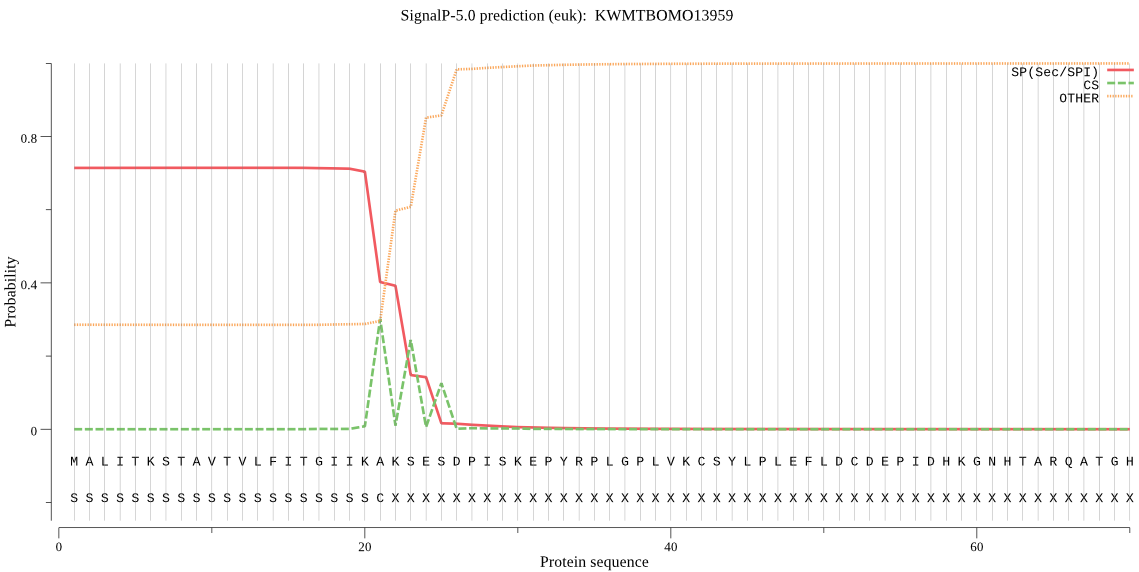
Length:
183
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.79258
Exp number, first 60 AAs:
0.639529999999999
Total prob of N-in:
0.27388
outside
1 - 113
TMhelix
114 - 133
inside
134 - 145
TMhelix
146 - 168
outside
169 - 183

Population Genetic Test Statistics
Pi
162.266337
Theta
140.307649
Tajima's D
0.366481
CLR
1.19332
CSRT
0.472276386180691
Interpretation
Uncertain
