Gene
KWMTBOMO13954
Pre Gene Modal
BGIBMGA011319
Annotation
PREDICTED:_integrator_complex_subunit_7_[Bombyx_mori]
Full name
Integrator complex subunit 7
Location in the cell
Mitochondrial Reliability : 1.641
Sequence
CDS
ATGATTGGTGTTAAAATGAATACATTCAACGACAATTCGGCGGAGCCTGAACAAGATGCAAATTCAGCATTGACCGAGCTCGATAAAGGCCTACGGTCGTGCAAAGTAGGAGAGCAATGTGAAGCCATCGTCCGGTTCCCTCGTCTTTTCGAAAAATATCCTTTCCCTATATTAATTAATTCATCTTTTCTTAAATTGTCTGACGTTTTTCGTATGGGTAACAATTTTTTGAGACTCTGGGTTTTGAGAGTCTGTCAACAAAGCGAAAAGCACCTAGATAAGATTCTGAACGTAGACGAGTTCTTAAGGAGGATCTTTAGTGTTCTACACTCAAACGATCCGGTGGCTAGGGCTCTCGCTCTGAGAACGCTTGGTGCAGTAGCAGGTATTATCCCAGAGCGCCAGAATGTACATCACGCAATCCGAAGGGGCTTGGATAGCCATGACAATGTTGAAGTTGATGCTGCTATATATGCTACTACTAGATTTGCTGCCCACTCCAATTCATTCGCAGTAGCCATGTGCAATAAGCTATCAGATATGGTGGAAAGTGAGGGCACTGCTGCTGAGAGACGAGCAAAACTTGTCAGAGCCCTGAGGACCGTTCATGGTGGAGCAATCAGAGCACAAGGAGTTTTGAAATTATTGAGATCATTGTTAGACAAGTTTCCTTCGTCAAGTTCGGTGCGAGCTTCAATCACTGCACTCACACATATAGCTTCAGATACAGTGGTTCATGTTCCTGATCAGGTTGAACTACTGCTAAATATAGCCATGAATGACGCCCGATCAGCCGTACGTCGTTCCGCACTGCTTGGCCTCAAGAAGTTGGCTGAGAATGCAGCTCTCTGGCCCGCACACTGTATACAGAACCTGGTCAGAGCCGCTAGCGATAGTCGAGACACTGACCACACTATGCTGTGCCTTCGAGTAATGCAGATTCTGGTGCAGTGCCCAGCAGTGTGCGCCACGGCAGGGTGTCCTCAGCGCGGCGAGTACAGTGCCCTGAGGCAGTACTGCAGTGAAGCCGCTCTCAGTGCTGACTTGAAGCAAGCCGCAACCGCTGCGGATGTCCTCACCAGGATCGTTGTTCATTGCTATGAAGAATGTCTTCCGGTGGAAGGTTCAGATCTGATGCTGGCTCTGGAGTCCCTGGTGATAGCGGCAGGGGCTGATAATGGGCAGAATATAAAGTCACTCAGGGTTGCGCTTAGGTGTTTGGTCCAGCTGAGCACTGCAGAACCCACAGTTTACGCTCCACGGACGGCATCGGTGCTTGCTGGACAACTGGCTACCAGTAGCTGGGCCAACCCTGGCCCGAGGGAGGCCTCGATCCTGGAAGCCCTGGGCGCGTTAGGTGGTGGAACCTACTGTCCCATGTCGGTGGCACCAGCGTTGCCCGCACTGCATCAAGCGTTGGAAGCCATGCAGAGAGAAAATGCGATAGCAAATCCGTCAGACGAAGGTAACGCTCTGGTGCTTCTCTGCACGGTGCTTTTCCAAGAACGGGCCGGGAAAGCGTTGACCTTTACCATCAGCAACGACTGGTGCACGAAGATCTTGGACGCGGTCCAAGTGGCTGACGGTTGGGTGCGATACAGAGTCGCCAGGGCCGCCTTGAGGTACGGGCATCACCACCTGGCCGCGGAGCTGCTGGTCCGGCTGGCGGCGCAGGCCCCCAGCGAGGCGGCTCAGCGCTGGCTGGCGGCGCTGCACCGGGCCGCCGCCGCCGACGCCAAGCTCGCCATCGACGGTAACGAGCTTATGTATATTATCCGTACACGCAGACAACTAGCGGACTAA
Protein
MIGVKMNTFNDNSAEPEQDANSALTELDKGLRSCKVGEQCEAIVRFPRLFEKYPFPILINSSFLKLSDVFRMGNNFLRLWVLRVCQQSEKHLDKILNVDEFLRRIFSVLHSNDPVARALALRTLGAVAGIIPERQNVHHAIRRGLDSHDNVEVDAAIYATTRFAAHSNSFAVAMCNKLSDMVESEGTAAERRAKLVRALRTVHGGAIRAQGVLKLLRSLLDKFPSSSSVRASITALTHIASDTVVHVPDQVELLLNIAMNDARSAVRRSALLGLKKLAENAALWPAHCIQNLVRAASDSRDTDHTMLCLRVMQILVQCPAVCATAGCPQRGEYSALRQYCSEAALSADLKQAATAADVLTRIVVHCYEECLPVEGSDLMLALESLVIAAGADNGQNIKSLRVALRCLVQLSTAEPTVYAPRTASVLAGQLATSSWANPGPREASILEALGALGGGTYCPMSVAPALPALHQALEAMQRENAIANPSDEGNALVLLCTVLFQERAGKALTFTISNDWCTKILDAVQVADGWVRYRVARAALRYGHHHLAAELLVRLAAQAPSEAAQRWLAALHRAAAADAKLAIDGNELMYIIRTRRQLAD
Summary
Description
Component of the Integrator complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. May play a role in DNA damage response (DDR) signaling during the S phase.
Subunit
Belongs to the multiprotein complex Integrator.
Similarity
Belongs to the Integrator subunit 7 family.
Keywords
Chromosome
Complete proteome
DNA damage
Nucleus
Reference proteome
Feature
chain Integrator complex subunit 7
Uniprot
H9JP65
A0A437BHS9
A0A2A4K3G7
A0A2A4K2F8
A0A2H1WHS8
L7X0U3
+ More
C3PPH1 A0A212FCZ7 A0A194RED4 A0A194QIC2 A0A2J7QT39 A0A1Y1NF75 A0A0L7QRY9 A0A0N0BFT0 E2ACI1 A0A151WNM2 A0A154P6S6 A0A088ALP4 A0A2A3ES99 A0A158N920 E0VNN0 E2BBN0 A0A0A9W714 A0A026W7V3 A0A146KV74 A0A151J7N6 N6TTR7 A0A195FU03 F4WHI4 T1H929 A0A151I6Y2 A0A195BVR4 A0A023EXU1 D6W9C5 A0A1B6HNP3 A0A139WMM3 A0A232F331 A0A1B6DJT3 K7J5H3 A0A0L0BTF0 A0A182HNR4 A0A1I8Q617 A0A182PUS5 A0A1I8NII0 A0A182SHY3 A0A1S4H6J2 Q7PMY5 A0A182LPS7 A0A182TQH5 A0A1Q3G3H8 A0A182YD83 A0A182KC86 A0A182XEB8 A0A182UWD8 A0A182Q2U1 A0A182RQT5 A0A182MBG1 A0A182MY85 A0A2G9P923 A0A182WG16 A0A336MZS9 A0A336MB09 J9JLK9 A0A1B0AR59 A0A084WB03 A0A1A9XGS7 W5JMI8 H0YY53 K7FYG8 U3KD91 A0A452HS34 A0A3L8SQV3 A0A151P3K7 A0A1U7SBD5 G1KFM6 A0A151P3Y6 A0A226MUR7 A0A151P3T9 G1NBV1 Q5ZL91 A0A226NY60 A0A0P5P4V4 A0A093QKI1 A0A218V3U7 A0A1A9ZG24 A0A091G180 D3DT96 A0A0A1WYV2 A0A2I0US64 A0A182FVR6
C3PPH1 A0A212FCZ7 A0A194RED4 A0A194QIC2 A0A2J7QT39 A0A1Y1NF75 A0A0L7QRY9 A0A0N0BFT0 E2ACI1 A0A151WNM2 A0A154P6S6 A0A088ALP4 A0A2A3ES99 A0A158N920 E0VNN0 E2BBN0 A0A0A9W714 A0A026W7V3 A0A146KV74 A0A151J7N6 N6TTR7 A0A195FU03 F4WHI4 T1H929 A0A151I6Y2 A0A195BVR4 A0A023EXU1 D6W9C5 A0A1B6HNP3 A0A139WMM3 A0A232F331 A0A1B6DJT3 K7J5H3 A0A0L0BTF0 A0A182HNR4 A0A1I8Q617 A0A182PUS5 A0A1I8NII0 A0A182SHY3 A0A1S4H6J2 Q7PMY5 A0A182LPS7 A0A182TQH5 A0A1Q3G3H8 A0A182YD83 A0A182KC86 A0A182XEB8 A0A182UWD8 A0A182Q2U1 A0A182RQT5 A0A182MBG1 A0A182MY85 A0A2G9P923 A0A182WG16 A0A336MZS9 A0A336MB09 J9JLK9 A0A1B0AR59 A0A084WB03 A0A1A9XGS7 W5JMI8 H0YY53 K7FYG8 U3KD91 A0A452HS34 A0A3L8SQV3 A0A151P3K7 A0A1U7SBD5 G1KFM6 A0A151P3Y6 A0A226MUR7 A0A151P3T9 G1NBV1 Q5ZL91 A0A226NY60 A0A0P5P4V4 A0A093QKI1 A0A218V3U7 A0A1A9ZG24 A0A091G180 D3DT96 A0A0A1WYV2 A0A2I0US64 A0A182FVR6
Pubmed
19121390
23674305
22118469
26354079
28004739
20798317
+ More
21347285 20566863 25401762 24508170 26823975 23537049 21719571 25474469 18362917 19820115 28648823 20075255 26108605 25315136 12364791 20966253 25244985 24438588 20920257 23761445 20360741 17381049 28562605 30282656 22293439 20838655 15642098 24621616 11181995 25830018
21347285 20566863 25401762 24508170 26823975 23537049 21719571 25474469 18362917 19820115 28648823 20075255 26108605 25315136 12364791 20966253 25244985 24438588 20920257 23761445 20360741 17381049 28562605 30282656 22293439 20838655 15642098 24621616 11181995 25830018
EMBL
BABH01009595
BABH01009596
RSAL01000054
RVE49984.1
NWSH01000204
PCG78444.1
+ More
PCG78445.1 ODYU01008749 SOQ52618.1 KC469894 AGC92734.1 CU681835 CAY54144.1 AGBW02009117 OWR51625.1 KQ460297 KPJ16193.1 KQ458761 KPJ05144.1 NEVH01011202 PNF31746.1 GEZM01005376 JAV96198.1 KQ414775 KOC61271.1 KQ435794 KOX73792.1 GL438528 EFN68847.1 KQ982907 KYQ49464.1 KQ434827 KZC07629.1 KZ288189 PBC34577.1 ADTU01009036 ADTU01009037 DS235341 EEB14986.1 GL447128 EFN86907.1 GBHO01039362 GBHO01039361 GBHO01039360 GBHO01039359 JAG04242.1 JAG04243.1 JAG04244.1 JAG04245.1 KK107372 EZA51721.1 GDHC01019104 GDHC01011811 JAP99524.1 JAQ06818.1 KQ979640 KYN20028.1 APGK01052548 KB741213 ENN72625.1 KQ981268 KYN43938.1 GL888161 EGI66338.1 ACPB03003465 KQ978451 KYM93935.1 KQ976405 KYM91886.1 GBBI01004896 JAC13816.1 KQ971312 EEZ98480.1 GECU01031402 JAS76304.1 KYB29182.1 NNAY01001170 OXU24898.1 GEDC01012572 GEDC01011357 JAS24726.1 JAS25941.1 JRES01001494 KNC22489.1 APCN01001971 AAAB01008966 EAA13050.5 GFDL01000687 JAV34358.1 AXCN02002037 AXCM01001977 KV922615 PIN99400.1 UFQT01003664 SSX35191.1 UFQT01000832 SSX27465.1 ABLF02025798 JXJN01002265 ATLV01022298 KE525331 KFB47397.1 ADMH02000621 ETN65592.1 ABQF01123290 AGCU01167073 AGCU01167074 AGCU01167075 AGCU01167076 AGTO01020510 QUSF01000008 RLW06890.1 AKHW03001146 KYO43638.1 KYO43639.1 MCFN01000449 OXB58749.1 KYO43640.1 AJ719843 AWGT02000284 OXB72434.1 GDIQ01133568 JAL18158.1 KL761277 KFW89066.1 MUZQ01000055 OWK60664.1 KL447600 KFO75106.1 CH471100 EAW93401.1 GBXI01010496 JAD03796.1 KZ505644 PKU48893.1
PCG78445.1 ODYU01008749 SOQ52618.1 KC469894 AGC92734.1 CU681835 CAY54144.1 AGBW02009117 OWR51625.1 KQ460297 KPJ16193.1 KQ458761 KPJ05144.1 NEVH01011202 PNF31746.1 GEZM01005376 JAV96198.1 KQ414775 KOC61271.1 KQ435794 KOX73792.1 GL438528 EFN68847.1 KQ982907 KYQ49464.1 KQ434827 KZC07629.1 KZ288189 PBC34577.1 ADTU01009036 ADTU01009037 DS235341 EEB14986.1 GL447128 EFN86907.1 GBHO01039362 GBHO01039361 GBHO01039360 GBHO01039359 JAG04242.1 JAG04243.1 JAG04244.1 JAG04245.1 KK107372 EZA51721.1 GDHC01019104 GDHC01011811 JAP99524.1 JAQ06818.1 KQ979640 KYN20028.1 APGK01052548 KB741213 ENN72625.1 KQ981268 KYN43938.1 GL888161 EGI66338.1 ACPB03003465 KQ978451 KYM93935.1 KQ976405 KYM91886.1 GBBI01004896 JAC13816.1 KQ971312 EEZ98480.1 GECU01031402 JAS76304.1 KYB29182.1 NNAY01001170 OXU24898.1 GEDC01012572 GEDC01011357 JAS24726.1 JAS25941.1 JRES01001494 KNC22489.1 APCN01001971 AAAB01008966 EAA13050.5 GFDL01000687 JAV34358.1 AXCN02002037 AXCM01001977 KV922615 PIN99400.1 UFQT01003664 SSX35191.1 UFQT01000832 SSX27465.1 ABLF02025798 JXJN01002265 ATLV01022298 KE525331 KFB47397.1 ADMH02000621 ETN65592.1 ABQF01123290 AGCU01167073 AGCU01167074 AGCU01167075 AGCU01167076 AGTO01020510 QUSF01000008 RLW06890.1 AKHW03001146 KYO43638.1 KYO43639.1 MCFN01000449 OXB58749.1 KYO43640.1 AJ719843 AWGT02000284 OXB72434.1 GDIQ01133568 JAL18158.1 KL761277 KFW89066.1 MUZQ01000055 OWK60664.1 KL447600 KFO75106.1 CH471100 EAW93401.1 GBXI01010496 JAD03796.1 KZ505644 PKU48893.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000235965 UP000053825 UP000053105 UP000000311 UP000075809 UP000076502 UP000005203 UP000242457 UP000005205 UP000009046 UP000008237 UP000053097 UP000078492 UP000019118 UP000078541 UP000007755 UP000015103 UP000078542 UP000078540 UP000007266 UP000215335 UP000002358 UP000037069 UP000075840 UP000095300 UP000075885 UP000095301 UP000075901 UP000007062 UP000075882 UP000075902 UP000076408 UP000075881 UP000076407 UP000075903 UP000075886 UP000075900 UP000075883 UP000075884 UP000075920 UP000007819 UP000092460 UP000030765 UP000092443 UP000000673 UP000007754 UP000007267 UP000016665 UP000291020 UP000276834 UP000050525 UP000189705 UP000001646 UP000198323 UP000001645 UP000000539 UP000198419 UP000053258 UP000197619 UP000092445 UP000053760 UP000069272
UP000235965 UP000053825 UP000053105 UP000000311 UP000075809 UP000076502 UP000005203 UP000242457 UP000005205 UP000009046 UP000008237 UP000053097 UP000078492 UP000019118 UP000078541 UP000007755 UP000015103 UP000078542 UP000078540 UP000007266 UP000215335 UP000002358 UP000037069 UP000075840 UP000095300 UP000075885 UP000095301 UP000075901 UP000007062 UP000075882 UP000075902 UP000076408 UP000075881 UP000076407 UP000075903 UP000075886 UP000075900 UP000075883 UP000075884 UP000075920 UP000007819 UP000092460 UP000030765 UP000092443 UP000000673 UP000007754 UP000007267 UP000016665 UP000291020 UP000276834 UP000050525 UP000189705 UP000001646 UP000198323 UP000001645 UP000000539 UP000198419 UP000053258 UP000197619 UP000092445 UP000053760 UP000069272
Pfam
PF00787 PX
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JP65
A0A437BHS9
A0A2A4K3G7
A0A2A4K2F8
A0A2H1WHS8
L7X0U3
+ More
C3PPH1 A0A212FCZ7 A0A194RED4 A0A194QIC2 A0A2J7QT39 A0A1Y1NF75 A0A0L7QRY9 A0A0N0BFT0 E2ACI1 A0A151WNM2 A0A154P6S6 A0A088ALP4 A0A2A3ES99 A0A158N920 E0VNN0 E2BBN0 A0A0A9W714 A0A026W7V3 A0A146KV74 A0A151J7N6 N6TTR7 A0A195FU03 F4WHI4 T1H929 A0A151I6Y2 A0A195BVR4 A0A023EXU1 D6W9C5 A0A1B6HNP3 A0A139WMM3 A0A232F331 A0A1B6DJT3 K7J5H3 A0A0L0BTF0 A0A182HNR4 A0A1I8Q617 A0A182PUS5 A0A1I8NII0 A0A182SHY3 A0A1S4H6J2 Q7PMY5 A0A182LPS7 A0A182TQH5 A0A1Q3G3H8 A0A182YD83 A0A182KC86 A0A182XEB8 A0A182UWD8 A0A182Q2U1 A0A182RQT5 A0A182MBG1 A0A182MY85 A0A2G9P923 A0A182WG16 A0A336MZS9 A0A336MB09 J9JLK9 A0A1B0AR59 A0A084WB03 A0A1A9XGS7 W5JMI8 H0YY53 K7FYG8 U3KD91 A0A452HS34 A0A3L8SQV3 A0A151P3K7 A0A1U7SBD5 G1KFM6 A0A151P3Y6 A0A226MUR7 A0A151P3T9 G1NBV1 Q5ZL91 A0A226NY60 A0A0P5P4V4 A0A093QKI1 A0A218V3U7 A0A1A9ZG24 A0A091G180 D3DT96 A0A0A1WYV2 A0A2I0US64 A0A182FVR6
C3PPH1 A0A212FCZ7 A0A194RED4 A0A194QIC2 A0A2J7QT39 A0A1Y1NF75 A0A0L7QRY9 A0A0N0BFT0 E2ACI1 A0A151WNM2 A0A154P6S6 A0A088ALP4 A0A2A3ES99 A0A158N920 E0VNN0 E2BBN0 A0A0A9W714 A0A026W7V3 A0A146KV74 A0A151J7N6 N6TTR7 A0A195FU03 F4WHI4 T1H929 A0A151I6Y2 A0A195BVR4 A0A023EXU1 D6W9C5 A0A1B6HNP3 A0A139WMM3 A0A232F331 A0A1B6DJT3 K7J5H3 A0A0L0BTF0 A0A182HNR4 A0A1I8Q617 A0A182PUS5 A0A1I8NII0 A0A182SHY3 A0A1S4H6J2 Q7PMY5 A0A182LPS7 A0A182TQH5 A0A1Q3G3H8 A0A182YD83 A0A182KC86 A0A182XEB8 A0A182UWD8 A0A182Q2U1 A0A182RQT5 A0A182MBG1 A0A182MY85 A0A2G9P923 A0A182WG16 A0A336MZS9 A0A336MB09 J9JLK9 A0A1B0AR59 A0A084WB03 A0A1A9XGS7 W5JMI8 H0YY53 K7FYG8 U3KD91 A0A452HS34 A0A3L8SQV3 A0A151P3K7 A0A1U7SBD5 G1KFM6 A0A151P3Y6 A0A226MUR7 A0A151P3T9 G1NBV1 Q5ZL91 A0A226NY60 A0A0P5P4V4 A0A093QKI1 A0A218V3U7 A0A1A9ZG24 A0A091G180 D3DT96 A0A0A1WYV2 A0A2I0US64 A0A182FVR6
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
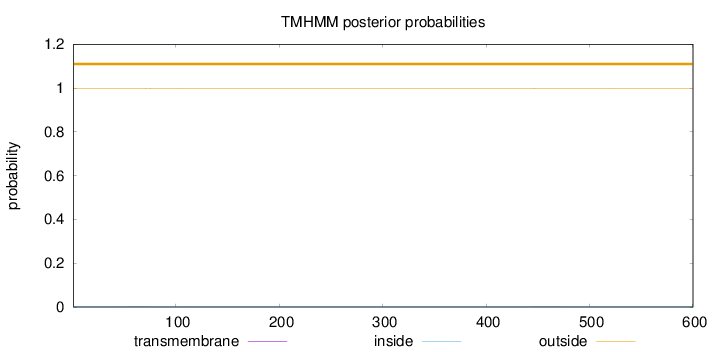
Length:
600
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0342
Exp number, first 60 AAs:
0.00389
Total prob of N-in:
0.00196
outside
1 - 600
Population Genetic Test Statistics
Pi
303.976618
Theta
203.47201
Tajima's D
1.447059
CLR
0.200689
CSRT
0.771761411929404
Interpretation
Uncertain
