Gene
KWMTBOMO13951
Pre Gene Modal
BGIBMGA011318
Annotation
PREDICTED:_adhesion_G_protein-coupled_receptor_A3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.063
Sequence
CDS
ATGTCGTTGCTATGCGCAAACGGGGTGCTCTCATACTGTCCCTCACTGTGTGTATGCAAAAGTAATAAGGCAGGCGAGGGGGCATCAGCGGAACCACTGCCGGGAGAACTCAAGCTCAAGTGCGGTGGAAGTCCAGCACCGATAACTGAACTTAAGGAAATCGACCTGAGCAAGCTGTGGACCATTGTAGTTAGTCTTAATTTATCCGGCAATGCTATTTCTACATTGTCGAGAGAGCTGTACTTGCCAAATCTACAGAAACTAGATTTAAGCAGAAACCAGATATCATTAATAGAATCGGATGCATTTTACAATATGACAGCTTTGCAAAAGTTAGACCTATCGCAGAACCACATCAGTAATGTGTACAAGGAAATGTTTAAAAGCTTAATCAATTTAGAGAGATTGATCCTGGCACAGAATCAGATATCTGTAATGGCATCGGGCACTTTTGATTATCTAGTTGCATTGAAGCATTTAGACATAACAGATAATCCTCTTATTTGCAACTGTGACTTACTTTGGGTGAGTGACTGGGTACGAATCACCAGGGTAAAACTTGTGGGTAATCCAAAATGTGCCTTCCCCGAGCACAAGACAAATACAACTATAAAAAAACTGAAGATATCATTAGATTTAAAAGCCTGCAGTAATGCTTTACCCATGACTTCATTGATCGCCAAGCCGAGCCACGATCAAGTAGTTTTCGAAGGGGATTCCTTGGTCTTGACTTGCAATGCACCATTCGCATCAGGCCTAATTAAATATACAGTTAAATGGGTGCACCCATTGCTTGAAATATGTGATGTTAATGTTACTTACACTGACATACAAGAGGAAGCAGTTGCTGAGACCACAATATATTTCTCGAATATAACTAACCATCATGCAGGAAACTGGACATGCATGTATAGTGACCAAAACAATGTGTACCACAACTACACCATAAATGTTTATGTGATATCAAATTCGACACAGTTCTGTGAGATTAAACACACAATGACAAATAAAGGATTGTACTCATGGCCACAGCTACTACTGAACCGCACATCTTCAGTACCGTGCCGCAGTGGAGAAGGCATGGCACACAGATATTGCCATGGGAATGGTACTTGGGGTGAAGTGGACACATCCGAATGTGCATACATAAGTAATGTGACGAAATTGCTTCAGCAGTTTGCACTGTTGAATGTGAGTTTAGTTCAGTATTCAGCAGTGAATGCTACGGAGAGACTGGCAATGTTGATACAGGAGAAGACATATCCTTTGGCTGAGATTACAGACCCAGATGATGTTATGTTCATAGCACGAGCAATTAGGAATTACATGCAGTATATTGCTGAAGAAAGAGATCTGGGTTCCGCATTATTAGATGTGATAAGTGCAGTGATGAACATATCAATGCCAGTTCTCGCCAAAGCCGAGACCAAGTACGGAGCATGCACAGATATGATGAATGCTGCAGAGGATATATCCGCTTATATTAACAATGTTCACGGACAAAAGTCAAATTTAGCGATAGAACGTTTTCGAGTGAGCGAGGGCTTCGGTGGAGCGACGTGCGTGTGGTACAGCGGGGGCGAAGTCCGAGGGGCAGAGCCTCCGAGACTGCACTGCACGGCCACCAACAGAACCGTTGCGCCCCTGTTCACACTATCCGACACGCTCATACATGCAACTGTGCAGATACCCCAATCTCTGATCTTCAGCACGACAGAATCTGGTGCCCTCGTTCAGATGAAGCCTTCGGACCTGGTGGTGTCGGTGTACGAGCACGCGGCGCTGTTCCCGCTGCTGCCGACCCTGGACGACGACGCCACGCTCACGGGCCACCGGTCGAAGGACTATTATGGCAGGAGCACCAAGCGGGAAGACATGAACATCGAGATCACCTCACCGGTTGTCGGATTTCAACTCAAGGGTACGGAGTGGTCGGGCGAGCTAGTGGAGCCAGTACTGGCGGGAGTGCGCGCTCGGCCCGGCGCTCGCACGCACGTAGCACTCTGGGACGCGCACACTCGCCACTGGACGCTTAACCATACAGATTGTCGCATATCGCACGCAGTGTCCGAGATGGTGATCATAAGCTGCAGCCGGCTGGCGCGCGTGGGGCTGGTGCAGAACGTGGAGACCGGCATCTACGGCGCCCGCACGGACGGCGCGCGCTTCAAGGTGTCGCACCCCGCCGTGTACGTGGGCAGTCTGCTGCTGGGCGGCTGCCTCGCCTGCGCCGCCATCACCTACGCCACGTGCCACGCCGACATACTGATGCCGAACAAAACCAAGCACGCCCTCGTCAACACCTGGGTGGCCATCTGTCTGCTCTGCTTCATGTACACCCTCGGCATCTACCAGACTGAGAACGTCAGGCTGTGCCAAGCGCTGGGACTGCTCATACACTACCTGTCGCTGTGCTGCTTGCTGTGGATGTGCGTGTCGGCCAGCAACATGTACAAGTGGGTGACGCGGGCCGACGGGCCGCCCAGGACGCCCGAGGACGAGCTACCGCCGGACGCGCCCGCGCACAAGCCCATACTGGGACTGTACCTGGTCGGCTGGGGCATAGCGCTCATAGTCTGCGGCATCTCCGGAGCCGTTAACATCAAGGACTACGCCGGCTACTCGCAGTGCTTCCTCAGCACGGCCCCGGCGCTGAGCGCGCTGTTCGTGCCCGGCGGCATCCTGTTGATGTTCCTGTTGGTGCTGTTCGCGCTCATACGCTGCACCATCCGGAACATGAACGTGCAGCTGTCGGAGGGCACGCAGGCCACCGAGAACGTCGACCTGGAGATGTGGGAGCCGCAGGTGCATGACCGCGCGGAGAGGAGGAGCGTGAAGTCGGCGCCGGACTCGGAGAGCGAGGACGCGGAGCACTCGCCCATCGTGCAGCTGCGGGCGCACGTGATCGTGGCGTGTCTGTACGTGGCGGTGTGGGCGTGCGGCGCCGTGGCCGTGTACCGGCCGCTGCCCGCCTACCTGCCGTACCAGGAGGACATCTGCAGCATCGTGTACGCCGTGTGCGCCACCGTGCTCGGCACCTTCGTGCTGTTCTTCTACGGCATAGCGCGCAGTGACGTCAGGGCGCAGTGGGCACTCGTGCATTGCTATTTTCAAAAAAGTAAACAGTGCTGCAGAAACAGGAGCGTGTTCGACGCGAACCAGCGCAACGTGCCCGCAGATCCCTCCGGAGCCGCCGCCCCCGCACCCCTGCCGAACGACAACCGGTCGCGGTCCGGCAGCCGGAGCTCCGGCAGGACCAACCCCAAGACCAACAACGGCGGCACGTACAAGGCGGGCGCTGAACTCAACGGGCAGACTCTACACAAGGACGTGTTGAAGAACACGAATGGAAAAACTCCGAACATAAACCTGGTTGTGCTGCACCGCCAACAGTACCGGTCCAACAACTCCATGCTCACGTACCCCGACGCCAGCACCGTGGCCCCCGATATATTCTACAACCCCAACCAGATGAATGTCGCCAAGAAGTTCTTCAAGAAGCAACGGCAGAACATGAAGAGGAACTGCCTTGAGTTGCCGACGAGAAATGACTACGACAACGACTCGCACTTCTCGCTGCCGCTGCCCACTAAGGAGGCGTACAGCGGGATGGCCAACATAATCAACTCTGGGTCCAAGGTCAACAATACGAACATACACGTTGAGCGGTCCAACAATGGCATCAAGGAGACCCGCAACGTCGCCAACCCGAACCTTCTGGAAGACGAACACAAGGACTACAAAAGCTACACGAACGATAAGAAGACTTGGAATAAAAACGAAGATCCGAAAACGGACAGAAGCCGAGTGATGAATATTTATACCAATGTGCCAGAGACTCAAGTGCCGCAGCATCAGGTCATCAAAGCGAACGCGCCCAGCAACCTCAATGCTCAGCGGAGCAGCGTATCGGAGGAGTGCCTGCAGAGTCACGCCGACCGCCCGGAGATGAGGACCATATCGCAGCAGTGCAGTCTGGAGTACAGCTCGGCGTCGGAGATAGCGATGCCGCACACCTGCTCCGACCAAACATTGAACACGCACTCGGAAATGACGTCACTGCAGGAAACGCACAGCGCTTTGTGCTCCACCAACGAGATCACCGACACGGAGAGCTTCGGCCAGTACATAGATTACATGCAGCCCAACAAGGGCCAAGACGGGAAGGAGAACAAGGACAAGTCGCTGCAGGACATATCGGAGGAATGCACCATGAATAGCGACGACGCCGCTCGCTCCGGGAGTCCGCAGCATCCGGACGCCGGGGAACTCGACAATCTGCTTCAGGATCCAGAGCTAGGGTCGAAAGATAAACTGGACGACGACAAAGACACGTTTTTCTTGGCGAAAACAACGTACGACTTCGAAACGTGCAGCACGAACGCGAGCGAACAGGGCTTCGAGAACGACAGTGACATTTACTGTCCCAACTACCAGATGTCCGAAGTGAGCATACGGAGCCACGGCCTGTACGCGCCCTCCCCCTCGTCCATGTGCCCAAACGAAATAAGCTTCAGCACAGAGGACGTTTCTAACATAGAGAACCAGTACGTCAACTACGATCCCGGATGGCGGAAAACGATAAAGAGCAATCCGAAATTGGGGAAGTACGTGCCCACTCCCGCGCTCAGCTGTCCCGATGTCAACAGGGACAGCAGCCCGGTGTCGTTCGCGAGCGAGATAGACGAACTGTACACTCAGATCACGAGTAGAGAGCGAGACAAGGACAGAATTAGTCGGAAGGACACCTGTTTAGATGTGACTTTGAACAGTGACGTCACGCTGCGGCCTGACAGTGACAGTTGTGCGAGCGAGGGCGAAGGTCGCGGGGAAACCGCCCTGTGA
Protein
MSLLCANGVLSYCPSLCVCKSNKAGEGASAEPLPGELKLKCGGSPAPITELKEIDLSKLWTIVVSLNLSGNAISTLSRELYLPNLQKLDLSRNQISLIESDAFYNMTALQKLDLSQNHISNVYKEMFKSLINLERLILAQNQISVMASGTFDYLVALKHLDITDNPLICNCDLLWVSDWVRITRVKLVGNPKCAFPEHKTNTTIKKLKISLDLKACSNALPMTSLIAKPSHDQVVFEGDSLVLTCNAPFASGLIKYTVKWVHPLLEICDVNVTYTDIQEEAVAETTIYFSNITNHHAGNWTCMYSDQNNVYHNYTINVYVISNSTQFCEIKHTMTNKGLYSWPQLLLNRTSSVPCRSGEGMAHRYCHGNGTWGEVDTSECAYISNVTKLLQQFALLNVSLVQYSAVNATERLAMLIQEKTYPLAEITDPDDVMFIARAIRNYMQYIAEERDLGSALLDVISAVMNISMPVLAKAETKYGACTDMMNAAEDISAYINNVHGQKSNLAIERFRVSEGFGGATCVWYSGGEVRGAEPPRLHCTATNRTVAPLFTLSDTLIHATVQIPQSLIFSTTESGALVQMKPSDLVVSVYEHAALFPLLPTLDDDATLTGHRSKDYYGRSTKREDMNIEITSPVVGFQLKGTEWSGELVEPVLAGVRARPGARTHVALWDAHTRHWTLNHTDCRISHAVSEMVIISCSRLARVGLVQNVETGIYGARTDGARFKVSHPAVYVGSLLLGGCLACAAITYATCHADILMPNKTKHALVNTWVAICLLCFMYTLGIYQTENVRLCQALGLLIHYLSLCCLLWMCVSASNMYKWVTRADGPPRTPEDELPPDAPAHKPILGLYLVGWGIALIVCGISGAVNIKDYAGYSQCFLSTAPALSALFVPGGILLMFLLVLFALIRCTIRNMNVQLSEGTQATENVDLEMWEPQVHDRAERRSVKSAPDSESEDAEHSPIVQLRAHVIVACLYVAVWACGAVAVYRPLPAYLPYQEDICSIVYAVCATVLGTFVLFFYGIARSDVRAQWALVHCYFQKSKQCCRNRSVFDANQRNVPADPSGAAAPAPLPNDNRSRSGSRSSGRTNPKTNNGGTYKAGAELNGQTLHKDVLKNTNGKTPNINLVVLHRQQYRSNNSMLTYPDASTVAPDIFYNPNQMNVAKKFFKKQRQNMKRNCLELPTRNDYDNDSHFSLPLPTKEAYSGMANIINSGSKVNNTNIHVERSNNGIKETRNVANPNLLEDEHKDYKSYTNDKKTWNKNEDPKTDRSRVMNIYTNVPETQVPQHQVIKANAPSNLNAQRSSVSEECLQSHADRPEMRTISQQCSLEYSSASEIAMPHTCSDQTLNTHSEMTSLQETHSALCSTNEITDTESFGQYIDYMQPNKGQDGKENKDKSLQDISEECTMNSDDAARSGSPQHPDAGELDNLLQDPELGSKDKLDDDKDTFFLAKTTYDFETCSTNASEQGFENDSDIYCPNYQMSEVSIRSHGLYAPSPSSMCPNEISFSTEDVSNIENQYVNYDPGWRKTIKSNPKLGKYVPTPALSCPDVNRDSSPVSFASEIDELYTQITSRERDKDRISRKDTCLDVTLNSDVTLRPDSDSCASEGEGRGETAL
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
H9JP64
A0A2A4K7V4
A0A2H1WT51
L7X1J2
A0A212FCZ8
C3PPF7
+ More
A0A194RFV9 A0A067RRK8 A0A026WQ39 A0A087ZNA8 A0A151IBS2 A0A310SU61 E9J2E9 A0A151X191 E2BSA8 A0A2A3E7X0 A0A151I134 F4WVS2 A0A158NMQ5 A0A195DBP7 A0A151JTR1 A0A1Y1K2Z1 A0A232FMS6 A0A0T6B3J2 A0A0C9RPD9 A0A0C9R2H7 E2ALM1 A0A0J7NWB4 A0A1W4XDJ2 A0A139WAE3 A0A1B0FY99 A0A0N0U386 A0A1B6D4N5 A0A1B0CY88 A0A336MNL9 A0A336MA78 A0A0K8V8M5 A0A034VYF2 W8BNX9 A0A1I8PYX6 T1IIR4 A0A2S2NN94 J9JJ88 A0A210Q2J9
A0A194RFV9 A0A067RRK8 A0A026WQ39 A0A087ZNA8 A0A151IBS2 A0A310SU61 E9J2E9 A0A151X191 E2BSA8 A0A2A3E7X0 A0A151I134 F4WVS2 A0A158NMQ5 A0A195DBP7 A0A151JTR1 A0A1Y1K2Z1 A0A232FMS6 A0A0T6B3J2 A0A0C9RPD9 A0A0C9R2H7 E2ALM1 A0A0J7NWB4 A0A1W4XDJ2 A0A139WAE3 A0A1B0FY99 A0A0N0U386 A0A1B6D4N5 A0A1B0CY88 A0A336MNL9 A0A336MA78 A0A0K8V8M5 A0A034VYF2 W8BNX9 A0A1I8PYX6 T1IIR4 A0A2S2NN94 J9JJ88 A0A210Q2J9
Pubmed
EMBL
BABH01009576
BABH01009577
NWSH01000057
PCG80096.1
ODYU01010475
SOQ55614.1
+ More
KC469894 AGC92731.1 AGBW02009117 OWR51621.1 CU467808 CAY54167.1 KQ460297 KPJ16190.1 KK852508 KDR22399.1 KK107152 EZA57219.1 KQ978078 KYM97173.1 KQ760287 OAD60879.1 GL767845 EFZ12993.1 KQ982588 KYQ54126.1 GL450157 EFN81421.1 KZ288357 PBC27151.1 KQ976593 KYM79651.1 GL888394 EGI61692.1 ADTU01020670 KQ981010 KYN10313.1 KQ981936 KYN32767.1 GEZM01098196 JAV53955.1 NNAY01000011 OXU32041.1 LJIG01016024 KRT81844.1 GBYB01010290 JAG80057.1 GBYB01010289 JAG80056.1 GL440609 EFN65630.1 LBMM01001227 KMQ96690.1 KQ971409 KYB24876.1 AJVK01000888 AJVK01000889 AJVK01000890 KQ435897 KOX69202.1 GEDC01016661 GEDC01009547 JAS20637.1 JAS27751.1 AJWK01035592 AJWK01035593 UFQS01000843 UFQT01000843 SSX07235.1 SSX27578.1 UFQT01000749 SSX26930.1 GDHF01017369 JAI34945.1 GAKP01010621 GAKP01010619 JAC48331.1 GAMC01003715 JAC02841.1 JH430212 GGMR01006026 MBY18645.1 ABLF02028785 NEDP02005205 OWF42899.1
KC469894 AGC92731.1 AGBW02009117 OWR51621.1 CU467808 CAY54167.1 KQ460297 KPJ16190.1 KK852508 KDR22399.1 KK107152 EZA57219.1 KQ978078 KYM97173.1 KQ760287 OAD60879.1 GL767845 EFZ12993.1 KQ982588 KYQ54126.1 GL450157 EFN81421.1 KZ288357 PBC27151.1 KQ976593 KYM79651.1 GL888394 EGI61692.1 ADTU01020670 KQ981010 KYN10313.1 KQ981936 KYN32767.1 GEZM01098196 JAV53955.1 NNAY01000011 OXU32041.1 LJIG01016024 KRT81844.1 GBYB01010290 JAG80057.1 GBYB01010289 JAG80056.1 GL440609 EFN65630.1 LBMM01001227 KMQ96690.1 KQ971409 KYB24876.1 AJVK01000888 AJVK01000889 AJVK01000890 KQ435897 KOX69202.1 GEDC01016661 GEDC01009547 JAS20637.1 JAS27751.1 AJWK01035592 AJWK01035593 UFQS01000843 UFQT01000843 SSX07235.1 SSX27578.1 UFQT01000749 SSX26930.1 GDHF01017369 JAI34945.1 GAKP01010621 GAKP01010619 JAC48331.1 GAMC01003715 JAC02841.1 JH430212 GGMR01006026 MBY18645.1 ABLF02028785 NEDP02005205 OWF42899.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000027135
UP000053097
+ More
UP000005203 UP000078542 UP000075809 UP000008237 UP000242457 UP000078540 UP000007755 UP000005205 UP000078492 UP000078541 UP000215335 UP000000311 UP000036403 UP000192223 UP000007266 UP000092462 UP000053105 UP000092461 UP000095300 UP000007819 UP000242188
UP000005203 UP000078542 UP000075809 UP000008237 UP000242457 UP000078540 UP000007755 UP000005205 UP000078492 UP000078541 UP000215335 UP000000311 UP000036403 UP000192223 UP000007266 UP000092462 UP000053105 UP000092461 UP000095300 UP000007819 UP000242188
PRIDE
Pfam
Interpro
IPR013783
Ig-like_fold
+ More
IPR003599 Ig_sub
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR036179 Ig-like_dom_sf
IPR036445 GPCR_2_extracell_dom_sf
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR001879 GPCR_2_extracellular_dom
IPR000203 GPS
IPR026906 LRR_5
IPR013766 Thioredoxin_domain
IPR000866 AhpC/TSA
IPR036249 Thioredoxin-like_sf
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR013151 Immunoglobulin
IPR003599 Ig_sub
IPR000483 Cys-rich_flank_reg_C
IPR007110 Ig-like_dom
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR036179 Ig-like_dom_sf
IPR036445 GPCR_2_extracell_dom_sf
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR001879 GPCR_2_extracellular_dom
IPR000203 GPS
IPR026906 LRR_5
IPR013766 Thioredoxin_domain
IPR000866 AhpC/TSA
IPR036249 Thioredoxin-like_sf
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR013151 Immunoglobulin
Gene 3D
ProteinModelPortal
H9JP64
A0A2A4K7V4
A0A2H1WT51
L7X1J2
A0A212FCZ8
C3PPF7
+ More
A0A194RFV9 A0A067RRK8 A0A026WQ39 A0A087ZNA8 A0A151IBS2 A0A310SU61 E9J2E9 A0A151X191 E2BSA8 A0A2A3E7X0 A0A151I134 F4WVS2 A0A158NMQ5 A0A195DBP7 A0A151JTR1 A0A1Y1K2Z1 A0A232FMS6 A0A0T6B3J2 A0A0C9RPD9 A0A0C9R2H7 E2ALM1 A0A0J7NWB4 A0A1W4XDJ2 A0A139WAE3 A0A1B0FY99 A0A0N0U386 A0A1B6D4N5 A0A1B0CY88 A0A336MNL9 A0A336MA78 A0A0K8V8M5 A0A034VYF2 W8BNX9 A0A1I8PYX6 T1IIR4 A0A2S2NN94 J9JJ88 A0A210Q2J9
A0A194RFV9 A0A067RRK8 A0A026WQ39 A0A087ZNA8 A0A151IBS2 A0A310SU61 E9J2E9 A0A151X191 E2BSA8 A0A2A3E7X0 A0A151I134 F4WVS2 A0A158NMQ5 A0A195DBP7 A0A151JTR1 A0A1Y1K2Z1 A0A232FMS6 A0A0T6B3J2 A0A0C9RPD9 A0A0C9R2H7 E2ALM1 A0A0J7NWB4 A0A1W4XDJ2 A0A139WAE3 A0A1B0FY99 A0A0N0U386 A0A1B6D4N5 A0A1B0CY88 A0A336MNL9 A0A336MA78 A0A0K8V8M5 A0A034VYF2 W8BNX9 A0A1I8PYX6 T1IIR4 A0A2S2NN94 J9JJ88 A0A210Q2J9
PDB
1W8A
E-value=5.88101e-18,
Score=229
Ontologies
GO
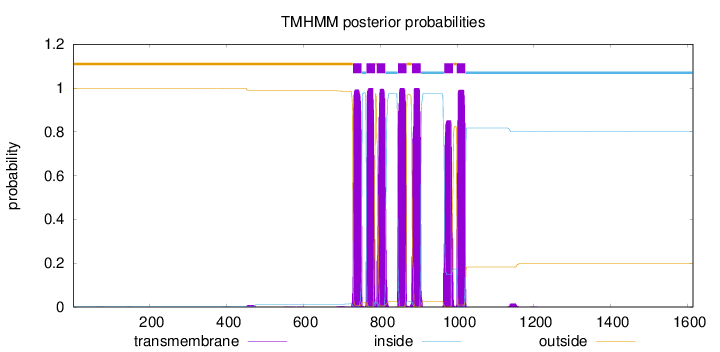
Topology
Length:
1614
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.60661
Exp number, first 60 AAs:
0.00093
Total prob of N-in:
0.00162
outside
1 - 728
TMhelix
729 - 751
inside
752 - 763
TMhelix
764 - 786
outside
787 - 790
TMhelix
791 - 813
inside
814 - 845
TMhelix
846 - 868
outside
869 - 882
TMhelix
883 - 905
inside
906 - 966
TMhelix
967 - 989
outside
990 - 998
TMhelix
999 - 1021
inside
1022 - 1614
Population Genetic Test Statistics
Pi
209.804428
Theta
212.770111
Tajima's D
-0.685993
CLR
1185.314103
CSRT
0.20213989300535
Interpretation
Uncertain
