Gene
KWMTBOMO13949
Pre Gene Modal
BGIBMGA011291
Annotation
PREDICTED:_cAMP_and_cAMP-inhibited_cGMP_3'?5'-cyclic_phosphodiesterase_10A-like_[Bombyx_mori]
Full name
Phosphodiesterase
Location in the cell
Nuclear Reliability : 2.468
Sequence
CDS
ATGATGCACGAACTCACTCCTAAGAATGTGAAATCGACAAAAAGCAAACAGGACGTGAAGCAGCAAGACGCCGAAGACGCTTTGTCCAAGAAGAGCTTGACCAAAGTACAATCTCACTTCTTGGTCCCGTCCGAGAAACTGGATAAGGTGACCAGACGAGTGATCGCTCCGTACGCGCAAATAGAACGGAAAGGCTTGAAAGAATCCGAACAGAAGGCCAGGAAATATCTCGATATTAACCGATTCAACGTGTTCCTGGATGACGAGATCGATTTGGACGCGTTATTGTACGAAACTGCCATGGTTTTGAAAAACTTTACCAATAGTTCCGGTAATTTAACTCGTTTGATGCTCCGTAGAGTCAATAAAGAAAACACCCTTTAA
Protein
MMHELTPKNVKSTKSKQDVKQQDAEDALSKKSLTKVQSHFLVPSEKLDKVTRRVIAPYAQIERKGLKESEQKARKYLDINRFNVFLDDEIDLDALLYETAMVLKNFTNSSGNLTRLMLRRVNKENTL
Summary
Cofactor
a divalent metal cation
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Uniprot
EC Number
3.1.4.-
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
Ontologies
PATHWAY
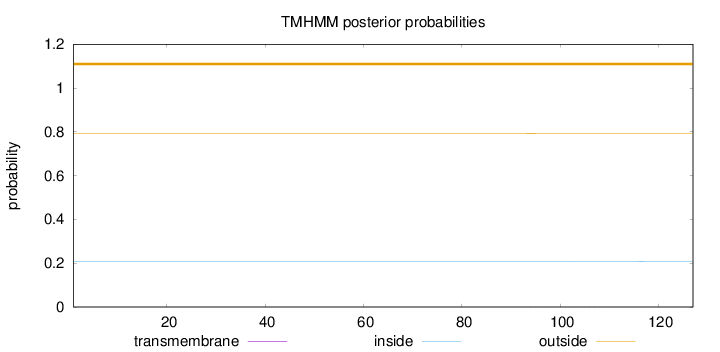
Topology
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00927
Exp number, first 60 AAs:
0
Total prob of N-in:
0.20784
outside
1 - 127
Population Genetic Test Statistics
Pi
21.900974
Theta
14.998334
Tajima's D
1.239944
CLR
0
CSRT
0.729763511824409
Interpretation
Uncertain
