Gene
KWMTBOMO13948
Pre Gene Modal
BGIBMGA011291
Annotation
PREDICTED:_cAMP_and_cAMP-inhibited_cGMP_3'?5'-cyclic_phosphodiesterase_10A-like_[Bombyx_mori]
Full name
Phosphodiesterase
Location in the cell
Cytoplasmic Reliability : 1.338 Extracellular Reliability : 1.753
Sequence
CDS
ATGAAGCTCCACCTGAAACCCTGTCGGCATGACGTCGAGGAACTGATGGAGACCAACGGCATTCTGCTGCCGCCTAGTAACTTCCGGACTTATGATTACGACATCAGCGAAGGGAACAGGGAGGAGATGGCGGGCCTGGTGTGCTACATGTTCAAGGAGACGTTTGCCGATCGTAATTTCGAGCGACAGCAGCTAGCGGAGTTCGTGTTATCCGTCCTCAGCTGTTACAGGGACAACCCGTACCACAACGCGCAGCACGCCTTCACGTTCACCCACACGATGTTCATGATATTAGTCAACAATACTGGCTACTTTGAGTTCGCTGAAGTGGCGGCCCTAATGATATCGGGTCTCTGTCACGACCTGGACCATCCCGGGTACAACAACAACTTCCTGATGCTCTGCAAGCACCCACTGGCCCTCATGTACAAGACGTCGATACTCGAGAATCATCATTATTTTCTCGCCAAGAAGATCGTTGAGGATAAACAAATATTCAGCAAGTTACCGATAGTCGATCAAGAACGAATATTGGAGGAGATGAAATACGCCATACTGTGCACCGACCTGGCGGTGTACTTCCAGCTGAGGGCGCAGCTGGCGCCGCTCATCGCCGACCAGACCTTCGATTGGAACGACAACCAGCACAGGAAACTACTCAAGGTACACTGCCGACCTTTTCTGCTCTTTGCGAAGTGA
Protein
MKLHLKPCRHDVEELMETNGILLPPSNFRTYDYDISEGNREEMAGLVCYMFKETFADRNFERQQLAEFVLSVLSCYRDNPYHNAQHAFTFTHTMFMILVNNTGYFEFAEVAALMISGLCHDLDHPGYNNNFLMLCKHPLALMYKTSILENHHYFLAKKIVEDKQIFSKLPIVDQERILEEMKYAILCTDLAVYFQLRAQLAPLIADQTFDWNDNQHRKLLKVHCRPFLLFAK
Summary
Cofactor
a divalent metal cation
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Uniprot
H9JP37
A0A212F6Z3
A0A2A4K8V9
A0A194QJR4
L7X1I8
A0A2H1W6A6
+ More
A0A0L7KN15 A0A139WM40 A0A1B6C6H2 A0A1B6CZB4 A0A1B6HGB4 A0A1B6GPX3 U4TTY5 U4UNK2 A0A1B6F756 A0A139WNG7 A0A1B6HPV1 A0A1B6LGV0 N6UE16 A0A067RGF4 B3RPP8 A0A1Y1KVY5 A0A1L8DL06 T1ID25 A0A0S7H156 A0A069DZ88 A0A224XC52 A0A2J7PWX3 A0A0L8I5L8 A0A1S3NDF0 A0A0P4VS35 A0A1S3NE72 A0A2R7WGW2 A0A0S7GXH9 A0A2P8Y691 A0A1B0CY76 M3ZTL5 A0A023F0G6 A0A1W4WN66 A0A1W4WYY5 A0A1W4WN57 A0A1S3Q7C1 A0A224X8R1 A0A3B5QMD8 A0A0S7H1T1 A0A3B5Q9T3 A0A0R4IJT4 A5PMD9 A0A0V0GAE4 F1QJC2 Q7ZW87 A0A3B5QYP5 A0A2J7PWY0 A0A0P4WRR5 W5K950 A0A2P4T170 A0A3B3RXT8 A0A3B3RX38 A7RVE3 A0A369SEH1 A0A3B3RXA8 T1HID7 A0A3B3XQY9 A0A087YDX5 A0A3B3XQY4 A0A3B3YY50 A0A3P9QJ34 A0A3P9QJZ9 A0A3P9QJ54 A0A2D0QKC9 B3RXL9 A0A315VRF0 A0A3B3V1V5 W6A375 A0A2D0QN85 A0A2D0QKM7 A0A2D0QKY9 A0A3B4DAI9 A0A2J7PWX5 K7E6Z8 A0A2U4CPT5 G3SP09 A0A3B4YNW7 A0A3Q1M418 A0A3B4YP55 F6URL1 K7FFY5 F1SBW6 A0A341CXC5 A0A1A8QT04 A0A2Y9M4C0 A0A1B6CTP1 R7UJM9 A0A2I2UWU4 A0A2Y9M3B8 A0A1Q3FNA8
A0A0L7KN15 A0A139WM40 A0A1B6C6H2 A0A1B6CZB4 A0A1B6HGB4 A0A1B6GPX3 U4TTY5 U4UNK2 A0A1B6F756 A0A139WNG7 A0A1B6HPV1 A0A1B6LGV0 N6UE16 A0A067RGF4 B3RPP8 A0A1Y1KVY5 A0A1L8DL06 T1ID25 A0A0S7H156 A0A069DZ88 A0A224XC52 A0A2J7PWX3 A0A0L8I5L8 A0A1S3NDF0 A0A0P4VS35 A0A1S3NE72 A0A2R7WGW2 A0A0S7GXH9 A0A2P8Y691 A0A1B0CY76 M3ZTL5 A0A023F0G6 A0A1W4WN66 A0A1W4WYY5 A0A1W4WN57 A0A1S3Q7C1 A0A224X8R1 A0A3B5QMD8 A0A0S7H1T1 A0A3B5Q9T3 A0A0R4IJT4 A5PMD9 A0A0V0GAE4 F1QJC2 Q7ZW87 A0A3B5QYP5 A0A2J7PWY0 A0A0P4WRR5 W5K950 A0A2P4T170 A0A3B3RXT8 A0A3B3RX38 A7RVE3 A0A369SEH1 A0A3B3RXA8 T1HID7 A0A3B3XQY9 A0A087YDX5 A0A3B3XQY4 A0A3B3YY50 A0A3P9QJ34 A0A3P9QJZ9 A0A3P9QJ54 A0A2D0QKC9 B3RXL9 A0A315VRF0 A0A3B3V1V5 W6A375 A0A2D0QN85 A0A2D0QKM7 A0A2D0QKY9 A0A3B4DAI9 A0A2J7PWX5 K7E6Z8 A0A2U4CPT5 G3SP09 A0A3B4YNW7 A0A3Q1M418 A0A3B4YP55 F6URL1 K7FFY5 F1SBW6 A0A341CXC5 A0A1A8QT04 A0A2Y9M4C0 A0A1B6CTP1 R7UJM9 A0A2I2UWU4 A0A2Y9M3B8 A0A1Q3FNA8
EC Number
3.1.4.-
Pubmed
EMBL
BABH01009575
AGBW02009949
OWR49512.1
NWSH01000057
PCG80102.1
KQ458761
+ More
KPJ05155.1 KC469894 AGC92726.1 ODYU01006306 SOQ48034.1 JTDY01008626 KOB64476.1 KQ971312 KYB29098.1 GEDC01028206 JAS09092.1 GEDC01018499 JAS18799.1 GECU01034004 JAS73702.1 GECZ01005310 JAS64459.1 KB630069 ERL83373.1 KB632376 ERL94053.1 GECZ01023769 JAS46000.1 KQ971311 KYB29482.1 GECU01031007 JAS76699.1 GEBQ01017056 JAT22921.1 APGK01031393 APGK01031394 APGK01031395 APGK01031396 APGK01031397 APGK01031398 APGK01031399 APGK01031400 APGK01031401 APGK01031402 KB740809 ENN78871.1 KK852480 KDR22951.1 DS985242 EDV27675.1 GEZM01075016 JAV64340.1 GFDF01006952 JAV07132.1 ACPB03013655 GBYX01448133 JAO33341.1 GBGD01001065 JAC87824.1 GFTR01006459 JAW09967.1 NEVH01020867 PNF20836.1 KQ416482 KOF96818.1 GDKW01001345 JAI55250.1 KK854805 PTY18892.1 GBYX01448122 JAO33352.1 PYGN01000878 PSN39765.1 AJWK01035539 GBBI01004268 JAC14444.1 GFTR01007679 JAW08747.1 GBYX01448127 JAO33347.1 BX323866 CABZ01035960 CR854838 GECL01001192 JAP04932.1 BC049532 AAH49532.1 PNF20837.1 GDRN01039842 JAI67178.1 PPHD01012981 POI30106.1 DS469543 EDO44626.1 NOWV01000017 RDD45317.1 ACPB03009364 ACPB03009365 AYCK01002113 DS985245 EDV24450.1 NHOQ01001229 PWA25693.1 JT405942 AHI50401.1 PNF20838.1 AGCU01201845 AGCU01201846 AGCU01201847 AGCU01201848 AGCU01201849 AGCU01201850 AEMK02000001 HAEG01014102 SBR96716.1 GEDC01020705 JAS16593.1 AMQN01007429 KB300677 ELU06420.1 AANG04000169 GFDL01005974 JAV29071.1
KPJ05155.1 KC469894 AGC92726.1 ODYU01006306 SOQ48034.1 JTDY01008626 KOB64476.1 KQ971312 KYB29098.1 GEDC01028206 JAS09092.1 GEDC01018499 JAS18799.1 GECU01034004 JAS73702.1 GECZ01005310 JAS64459.1 KB630069 ERL83373.1 KB632376 ERL94053.1 GECZ01023769 JAS46000.1 KQ971311 KYB29482.1 GECU01031007 JAS76699.1 GEBQ01017056 JAT22921.1 APGK01031393 APGK01031394 APGK01031395 APGK01031396 APGK01031397 APGK01031398 APGK01031399 APGK01031400 APGK01031401 APGK01031402 KB740809 ENN78871.1 KK852480 KDR22951.1 DS985242 EDV27675.1 GEZM01075016 JAV64340.1 GFDF01006952 JAV07132.1 ACPB03013655 GBYX01448133 JAO33341.1 GBGD01001065 JAC87824.1 GFTR01006459 JAW09967.1 NEVH01020867 PNF20836.1 KQ416482 KOF96818.1 GDKW01001345 JAI55250.1 KK854805 PTY18892.1 GBYX01448122 JAO33352.1 PYGN01000878 PSN39765.1 AJWK01035539 GBBI01004268 JAC14444.1 GFTR01007679 JAW08747.1 GBYX01448127 JAO33347.1 BX323866 CABZ01035960 CR854838 GECL01001192 JAP04932.1 BC049532 AAH49532.1 PNF20837.1 GDRN01039842 JAI67178.1 PPHD01012981 POI30106.1 DS469543 EDO44626.1 NOWV01000017 RDD45317.1 ACPB03009364 ACPB03009365 AYCK01002113 DS985245 EDV24450.1 NHOQ01001229 PWA25693.1 JT405942 AHI50401.1 PNF20838.1 AGCU01201845 AGCU01201846 AGCU01201847 AGCU01201848 AGCU01201849 AGCU01201850 AEMK02000001 HAEG01014102 SBR96716.1 GEDC01020705 JAS16593.1 AMQN01007429 KB300677 ELU06420.1 AANG04000169 GFDL01005974 JAV29071.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000037510
UP000007266
+ More
UP000030742 UP000019118 UP000027135 UP000009022 UP000015103 UP000235965 UP000053454 UP000087266 UP000245037 UP000092461 UP000002852 UP000192223 UP000000437 UP000018467 UP000261540 UP000001593 UP000253843 UP000261480 UP000028760 UP000242638 UP000221080 UP000261500 UP000261440 UP000002279 UP000245320 UP000007646 UP000261360 UP000009136 UP000007267 UP000008227 UP000252040 UP000248483 UP000014760 UP000011712
UP000030742 UP000019118 UP000027135 UP000009022 UP000015103 UP000235965 UP000053454 UP000087266 UP000245037 UP000092461 UP000002852 UP000192223 UP000000437 UP000018467 UP000261540 UP000001593 UP000253843 UP000261480 UP000028760 UP000242638 UP000221080 UP000261500 UP000261440 UP000002279 UP000245320 UP000007646 UP000261360 UP000009136 UP000007267 UP000008227 UP000252040 UP000248483 UP000014760 UP000011712
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JP37
A0A212F6Z3
A0A2A4K8V9
A0A194QJR4
L7X1I8
A0A2H1W6A6
+ More
A0A0L7KN15 A0A139WM40 A0A1B6C6H2 A0A1B6CZB4 A0A1B6HGB4 A0A1B6GPX3 U4TTY5 U4UNK2 A0A1B6F756 A0A139WNG7 A0A1B6HPV1 A0A1B6LGV0 N6UE16 A0A067RGF4 B3RPP8 A0A1Y1KVY5 A0A1L8DL06 T1ID25 A0A0S7H156 A0A069DZ88 A0A224XC52 A0A2J7PWX3 A0A0L8I5L8 A0A1S3NDF0 A0A0P4VS35 A0A1S3NE72 A0A2R7WGW2 A0A0S7GXH9 A0A2P8Y691 A0A1B0CY76 M3ZTL5 A0A023F0G6 A0A1W4WN66 A0A1W4WYY5 A0A1W4WN57 A0A1S3Q7C1 A0A224X8R1 A0A3B5QMD8 A0A0S7H1T1 A0A3B5Q9T3 A0A0R4IJT4 A5PMD9 A0A0V0GAE4 F1QJC2 Q7ZW87 A0A3B5QYP5 A0A2J7PWY0 A0A0P4WRR5 W5K950 A0A2P4T170 A0A3B3RXT8 A0A3B3RX38 A7RVE3 A0A369SEH1 A0A3B3RXA8 T1HID7 A0A3B3XQY9 A0A087YDX5 A0A3B3XQY4 A0A3B3YY50 A0A3P9QJ34 A0A3P9QJZ9 A0A3P9QJ54 A0A2D0QKC9 B3RXL9 A0A315VRF0 A0A3B3V1V5 W6A375 A0A2D0QN85 A0A2D0QKM7 A0A2D0QKY9 A0A3B4DAI9 A0A2J7PWX5 K7E6Z8 A0A2U4CPT5 G3SP09 A0A3B4YNW7 A0A3Q1M418 A0A3B4YP55 F6URL1 K7FFY5 F1SBW6 A0A341CXC5 A0A1A8QT04 A0A2Y9M4C0 A0A1B6CTP1 R7UJM9 A0A2I2UWU4 A0A2Y9M3B8 A0A1Q3FNA8
A0A0L7KN15 A0A139WM40 A0A1B6C6H2 A0A1B6CZB4 A0A1B6HGB4 A0A1B6GPX3 U4TTY5 U4UNK2 A0A1B6F756 A0A139WNG7 A0A1B6HPV1 A0A1B6LGV0 N6UE16 A0A067RGF4 B3RPP8 A0A1Y1KVY5 A0A1L8DL06 T1ID25 A0A0S7H156 A0A069DZ88 A0A224XC52 A0A2J7PWX3 A0A0L8I5L8 A0A1S3NDF0 A0A0P4VS35 A0A1S3NE72 A0A2R7WGW2 A0A0S7GXH9 A0A2P8Y691 A0A1B0CY76 M3ZTL5 A0A023F0G6 A0A1W4WN66 A0A1W4WYY5 A0A1W4WN57 A0A1S3Q7C1 A0A224X8R1 A0A3B5QMD8 A0A0S7H1T1 A0A3B5Q9T3 A0A0R4IJT4 A5PMD9 A0A0V0GAE4 F1QJC2 Q7ZW87 A0A3B5QYP5 A0A2J7PWY0 A0A0P4WRR5 W5K950 A0A2P4T170 A0A3B3RXT8 A0A3B3RX38 A7RVE3 A0A369SEH1 A0A3B3RXA8 T1HID7 A0A3B3XQY9 A0A087YDX5 A0A3B3XQY4 A0A3B3YY50 A0A3P9QJ34 A0A3P9QJZ9 A0A3P9QJ54 A0A2D0QKC9 B3RXL9 A0A315VRF0 A0A3B3V1V5 W6A375 A0A2D0QN85 A0A2D0QKM7 A0A2D0QKY9 A0A3B4DAI9 A0A2J7PWX5 K7E6Z8 A0A2U4CPT5 G3SP09 A0A3B4YNW7 A0A3Q1M418 A0A3B4YP55 F6URL1 K7FFY5 F1SBW6 A0A341CXC5 A0A1A8QT04 A0A2Y9M4C0 A0A1B6CTP1 R7UJM9 A0A2I2UWU4 A0A2Y9M3B8 A0A1Q3FNA8
PDB
3HR1
E-value=3.44835e-33,
Score=351
Ontologies
GO
Topology
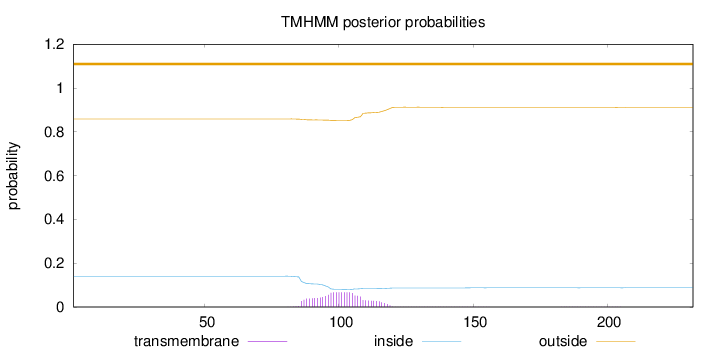
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.51245
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14086
outside
1 - 232
Population Genetic Test Statistics
Pi
243.580533
Theta
175.755476
Tajima's D
1.757767
CLR
0.384573
CSRT
0.84000799960002
Interpretation
Uncertain
