Pre Gene Modal
BGIBMGA011317
Annotation
PREDICTED:_sorting_nexin-12_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.555 Nuclear Reliability : 1.74
Sequence
CDS
ATGATGGCTGAGGATGCAACAGCCGAAGCTACTAGACGTCTCAACGTTAAGAAACAAACGTTGGATGACGCGTACGCGGCGCCGGCGAATTTCTTAGAAATCGACGTTCTTAATCCAGTGACCACTATGGGTGTTGGTAAAAAACGCTACACGGATTACGAAGTTCGTATGAGGACAAACCTACCCGTTTTCAAAGTGAAAGATTCGAGCGTCAGACGGCGGTATAGCGACTTCGAGTGGCTCAGGAATGAATTGGAGAGGGACAGCAAGATAGTCGTACCTCCCTTGCCCGGCAAAGCTCTGAAGAGACAGTTGCCGTTCAGAGGCGATGATGGTATATTTGAGGAGGAATTTATAGAGGACCGCAGAAAAGGTCTGGAAGTGTTCATCAACAAGATAGCCGGGCACCCGCTGGCGCAGAACGAGCGCTGCCTCCACATGTTCCTGCAGGAGCCCACCATCGACAGGAGCTACGTGCCCGGCAAGATCAGGAACACATAA
Protein
MMAEDATAEATRRLNVKKQTLDDAYAAPANFLEIDVLNPVTTMGVGKKRYTDYEVRMRTNLPVFKVKDSSVRRRYSDFEWLRNELERDSKIVVPPLPGKALKRQLPFRGDDGIFEEEFIEDRRKGLEVFINKIAGHPLAQNERCLHMFLQEPTIDRSYVPGKIRNT
Summary
Uniprot
H9JP63
A0A2H1WRC4
A0A2A4K8H3
A0A194QHX0
A0A1E1W0Z6
L7X1S1
+ More
D6W8L6 C3PPF4 A0A1Y1LWG3 A0A212FD42 A0A182H1V3 B4NHZ7 U4U778 J3JTF2 A0A1W4WMN4 A0A1L8DVW6 B4JGS1 T1E893 A0A2M3Z838 A0A2M4ANV4 A0A2M4C1J1 W5JSZ8 A0A084WSR3 B4QS88 A0A3B0K3P0 Q29BS2 B4GN06 B4PPC3 B3M1X6 B4HGV6 B3P491 Q9VG51 A0A0T6B412 A0A182F8L7 A0A1W4UN59 A0A0K8VXL3 A0A0A1WZP1 A0A034VX97 B4MC73 B4K7G6 B0W379 A0A0A9X9Y5 A0A182QFF7 A0A0M4F4A9 A0A0K8TQE9 A0A1Q3FDW6 A0A182M7V5 A0A182VHS7 A0A182JZF3 A0A182TW52 A0A182VSA8 A0A182XK53 A0A182KX08 Q7QDI2 A0A182R830 A0A182HJT7 A0A182PMX9 U5EI23 T1PC58 A0A151IRM0 A0A195BWJ8 A0A195FQJ7 A0A158NUW6 E1ZWA0 A0A151WT21 A0A2H8TFU9 C4WTD1 A0A0L7R8J5 A0A0L0CGE8 A0A195CRM6 A0A224XV01 A0A069DPS7 R4G544 Q17CA9 E2B7F6 T1D517 A0A1L8EDS5 A0A026W2F6 A0A2A3E517 V9IKW3 A0A0N0BC78 A0A088A6E1 K7IX60 A0A1B0G8S6 A0A1A9XJ17 A0A1A9V744 A0A1B0BXY8 A0A1A9W601 A0A1S3D9U8 A0A336MJL0 A0A154PJ85 A0A067RFT6 A0A1I8PUQ9 A0A2J7PE96 A0A0P4VY86 A0A023FAP0 A0A1B6GNE2 A0A1B6JV27 A0A182ITT6 A0A1B6DHG3
D6W8L6 C3PPF4 A0A1Y1LWG3 A0A212FD42 A0A182H1V3 B4NHZ7 U4U778 J3JTF2 A0A1W4WMN4 A0A1L8DVW6 B4JGS1 T1E893 A0A2M3Z838 A0A2M4ANV4 A0A2M4C1J1 W5JSZ8 A0A084WSR3 B4QS88 A0A3B0K3P0 Q29BS2 B4GN06 B4PPC3 B3M1X6 B4HGV6 B3P491 Q9VG51 A0A0T6B412 A0A182F8L7 A0A1W4UN59 A0A0K8VXL3 A0A0A1WZP1 A0A034VX97 B4MC73 B4K7G6 B0W379 A0A0A9X9Y5 A0A182QFF7 A0A0M4F4A9 A0A0K8TQE9 A0A1Q3FDW6 A0A182M7V5 A0A182VHS7 A0A182JZF3 A0A182TW52 A0A182VSA8 A0A182XK53 A0A182KX08 Q7QDI2 A0A182R830 A0A182HJT7 A0A182PMX9 U5EI23 T1PC58 A0A151IRM0 A0A195BWJ8 A0A195FQJ7 A0A158NUW6 E1ZWA0 A0A151WT21 A0A2H8TFU9 C4WTD1 A0A0L7R8J5 A0A0L0CGE8 A0A195CRM6 A0A224XV01 A0A069DPS7 R4G544 Q17CA9 E2B7F6 T1D517 A0A1L8EDS5 A0A026W2F6 A0A2A3E517 V9IKW3 A0A0N0BC78 A0A088A6E1 K7IX60 A0A1B0G8S6 A0A1A9XJ17 A0A1A9V744 A0A1B0BXY8 A0A1A9W601 A0A1S3D9U8 A0A336MJL0 A0A154PJ85 A0A067RFT6 A0A1I8PUQ9 A0A2J7PE96 A0A0P4VY86 A0A023FAP0 A0A1B6GNE2 A0A1B6JV27 A0A182ITT6 A0A1B6DHG3
Pubmed
19121390
26354079
23674305
18362917
19820115
28004739
+ More
22118469 26483478 17994087 23537049 22516182 20920257 23761445 24438588 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 25401762 26823975 26369729 20966253 12364791 14747013 17210077 25315136 21347285 20798317 26108605 26334808 17510324 24330624 24508170 30249741 20075255 24845553 27129103 25474469
22118469 26483478 17994087 23537049 22516182 20920257 23761445 24438588 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 25401762 26823975 26369729 20966253 12364791 14747013 17210077 25315136 21347285 20798317 26108605 26334808 17510324 24330624 24508170 30249741 20075255 24845553 27129103 25474469
EMBL
BABH01009575
ODYU01010475
SOQ55611.1
NWSH01000057
PCG80103.1
KQ458761
+ More
KPJ05153.1 GDQN01010408 JAT80646.1 KC469894 AGC92728.1 KQ971312 EEZ98290.1 CU467808 CAY54164.1 GEZM01045194 JAV77869.1 AGBW02009117 OWR51618.1 JXUM01023259 JXUM01023260 JXUM01023261 JXUM01023262 KQ560656 KXJ81412.1 CH964272 EDW83647.1 KB631815 ERL86456.1 BT126508 AEE61472.1 GFDF01003525 JAV10559.1 CH916369 EDV92675.1 GAMD01002377 JAA99213.1 GGFM01003933 MBW24684.1 GGFK01009132 MBW42453.1 GGFJ01010028 MBW59169.1 ADMH02000563 GGFL01004312 ETN65879.1 MBW68490.1 ATLV01026697 KE525417 KFB53257.1 CM000364 EDX13184.1 OUUW01000007 SPP82560.1 CM000070 EAL26925.1 CH479186 EDW39132.1 CM000160 EDW97129.1 CH902617 EDV42236.1 CH480815 EDW42427.1 CH954181 EDV49336.1 AE014297 AY071431 AAF54838.1 AAL49053.1 AAN13550.2 LJIG01009898 KRT82160.1 GDHF01026186 GDHF01020641 GDHF01016552 GDHF01016354 GDHF01008715 JAI26128.1 JAI31673.1 JAI35762.1 JAI35960.1 JAI43599.1 GBXI01011331 GBXI01010449 JAD02961.1 JAD03843.1 GAKP01011885 GAKP01011884 GAKP01011883 JAC47069.1 CH940656 EDW58694.1 CH933806 EDW15310.1 DS231831 EDS31222.1 GBHO01026795 GBRD01013524 GBRD01013523 GDHC01014314 JAG16809.1 JAG52302.1 JAQ04315.1 AXCN02000110 CP012526 ALC46170.1 GDAI01001498 JAI16105.1 GFDL01009285 JAV25760.1 AXCM01001279 AAAB01008851 EAA07353.4 APCN01002209 GANO01002799 JAB57072.1 KA645705 AFP60334.1 KQ981122 KYN09346.1 KQ976401 KYM92655.1 KQ981335 KYN42572.1 ADTU01026790 ADTU01026791 GL434781 EFN74540.1 KQ982769 KYQ50937.1 GFXV01001188 MBW12993.1 ABLF02034746 AK340566 BAH71151.1 KQ414632 KOC67207.1 JRES01000413 KNC31468.1 KQ977444 KYN02764.1 GFTR01002768 JAW13658.1 GBGD01003257 JAC85632.1 ACPB03009364 GAHY01000810 JAA76700.1 CH477310 EAT43966.1 GL446181 EFN88331.1 GALA01000733 JAA94119.1 GFDG01001967 JAV16832.1 KK107467 QOIP01000013 EZA50262.1 RLU15544.1 KZ288405 PBC26171.1 JR052353 AEY61680.1 KQ435922 KOX68538.1 AAZX01000605 AAZX01008772 CCAG010009134 JXJN01022426 UFQT01001447 UFQT01002076 SSX30604.1 SSX32589.1 KQ434936 KZC11905.1 KK852498 KDR22612.1 NEVH01026106 PNF14663.1 GDKW01001010 JAI55585.1 GBBI01000608 JAC18104.1 GECZ01005844 JAS63925.1 GECU01004943 JAT02764.1 GEDC01012159 JAS25139.1
KPJ05153.1 GDQN01010408 JAT80646.1 KC469894 AGC92728.1 KQ971312 EEZ98290.1 CU467808 CAY54164.1 GEZM01045194 JAV77869.1 AGBW02009117 OWR51618.1 JXUM01023259 JXUM01023260 JXUM01023261 JXUM01023262 KQ560656 KXJ81412.1 CH964272 EDW83647.1 KB631815 ERL86456.1 BT126508 AEE61472.1 GFDF01003525 JAV10559.1 CH916369 EDV92675.1 GAMD01002377 JAA99213.1 GGFM01003933 MBW24684.1 GGFK01009132 MBW42453.1 GGFJ01010028 MBW59169.1 ADMH02000563 GGFL01004312 ETN65879.1 MBW68490.1 ATLV01026697 KE525417 KFB53257.1 CM000364 EDX13184.1 OUUW01000007 SPP82560.1 CM000070 EAL26925.1 CH479186 EDW39132.1 CM000160 EDW97129.1 CH902617 EDV42236.1 CH480815 EDW42427.1 CH954181 EDV49336.1 AE014297 AY071431 AAF54838.1 AAL49053.1 AAN13550.2 LJIG01009898 KRT82160.1 GDHF01026186 GDHF01020641 GDHF01016552 GDHF01016354 GDHF01008715 JAI26128.1 JAI31673.1 JAI35762.1 JAI35960.1 JAI43599.1 GBXI01011331 GBXI01010449 JAD02961.1 JAD03843.1 GAKP01011885 GAKP01011884 GAKP01011883 JAC47069.1 CH940656 EDW58694.1 CH933806 EDW15310.1 DS231831 EDS31222.1 GBHO01026795 GBRD01013524 GBRD01013523 GDHC01014314 JAG16809.1 JAG52302.1 JAQ04315.1 AXCN02000110 CP012526 ALC46170.1 GDAI01001498 JAI16105.1 GFDL01009285 JAV25760.1 AXCM01001279 AAAB01008851 EAA07353.4 APCN01002209 GANO01002799 JAB57072.1 KA645705 AFP60334.1 KQ981122 KYN09346.1 KQ976401 KYM92655.1 KQ981335 KYN42572.1 ADTU01026790 ADTU01026791 GL434781 EFN74540.1 KQ982769 KYQ50937.1 GFXV01001188 MBW12993.1 ABLF02034746 AK340566 BAH71151.1 KQ414632 KOC67207.1 JRES01000413 KNC31468.1 KQ977444 KYN02764.1 GFTR01002768 JAW13658.1 GBGD01003257 JAC85632.1 ACPB03009364 GAHY01000810 JAA76700.1 CH477310 EAT43966.1 GL446181 EFN88331.1 GALA01000733 JAA94119.1 GFDG01001967 JAV16832.1 KK107467 QOIP01000013 EZA50262.1 RLU15544.1 KZ288405 PBC26171.1 JR052353 AEY61680.1 KQ435922 KOX68538.1 AAZX01000605 AAZX01008772 CCAG010009134 JXJN01022426 UFQT01001447 UFQT01002076 SSX30604.1 SSX32589.1 KQ434936 KZC11905.1 KK852498 KDR22612.1 NEVH01026106 PNF14663.1 GDKW01001010 JAI55585.1 GBBI01000608 JAC18104.1 GECZ01005844 JAS63925.1 GECU01004943 JAT02764.1 GEDC01012159 JAS25139.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007266
UP000007151
UP000069940
+ More
UP000249989 UP000007798 UP000030742 UP000192223 UP000001070 UP000000673 UP000030765 UP000000304 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000803 UP000069272 UP000192221 UP000008792 UP000009192 UP000002320 UP000075886 UP000092553 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000075885 UP000095301 UP000078492 UP000078540 UP000078541 UP000005205 UP000000311 UP000075809 UP000007819 UP000053825 UP000037069 UP000078542 UP000015103 UP000008820 UP000008237 UP000053097 UP000279307 UP000242457 UP000053105 UP000005203 UP000002358 UP000092444 UP000092443 UP000078200 UP000092460 UP000091820 UP000079169 UP000076502 UP000027135 UP000095300 UP000235965 UP000075880
UP000249989 UP000007798 UP000030742 UP000192223 UP000001070 UP000000673 UP000030765 UP000000304 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000803 UP000069272 UP000192221 UP000008792 UP000009192 UP000002320 UP000075886 UP000092553 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000075885 UP000095301 UP000078492 UP000078540 UP000078541 UP000005205 UP000000311 UP000075809 UP000007819 UP000053825 UP000037069 UP000078542 UP000015103 UP000008820 UP000008237 UP000053097 UP000279307 UP000242457 UP000053105 UP000005203 UP000002358 UP000092444 UP000092443 UP000078200 UP000092460 UP000091820 UP000079169 UP000076502 UP000027135 UP000095300 UP000235965 UP000075880
SUPFAM
SSF64268
SSF64268
Gene 3D
ProteinModelPortal
H9JP63
A0A2H1WRC4
A0A2A4K8H3
A0A194QHX0
A0A1E1W0Z6
L7X1S1
+ More
D6W8L6 C3PPF4 A0A1Y1LWG3 A0A212FD42 A0A182H1V3 B4NHZ7 U4U778 J3JTF2 A0A1W4WMN4 A0A1L8DVW6 B4JGS1 T1E893 A0A2M3Z838 A0A2M4ANV4 A0A2M4C1J1 W5JSZ8 A0A084WSR3 B4QS88 A0A3B0K3P0 Q29BS2 B4GN06 B4PPC3 B3M1X6 B4HGV6 B3P491 Q9VG51 A0A0T6B412 A0A182F8L7 A0A1W4UN59 A0A0K8VXL3 A0A0A1WZP1 A0A034VX97 B4MC73 B4K7G6 B0W379 A0A0A9X9Y5 A0A182QFF7 A0A0M4F4A9 A0A0K8TQE9 A0A1Q3FDW6 A0A182M7V5 A0A182VHS7 A0A182JZF3 A0A182TW52 A0A182VSA8 A0A182XK53 A0A182KX08 Q7QDI2 A0A182R830 A0A182HJT7 A0A182PMX9 U5EI23 T1PC58 A0A151IRM0 A0A195BWJ8 A0A195FQJ7 A0A158NUW6 E1ZWA0 A0A151WT21 A0A2H8TFU9 C4WTD1 A0A0L7R8J5 A0A0L0CGE8 A0A195CRM6 A0A224XV01 A0A069DPS7 R4G544 Q17CA9 E2B7F6 T1D517 A0A1L8EDS5 A0A026W2F6 A0A2A3E517 V9IKW3 A0A0N0BC78 A0A088A6E1 K7IX60 A0A1B0G8S6 A0A1A9XJ17 A0A1A9V744 A0A1B0BXY8 A0A1A9W601 A0A1S3D9U8 A0A336MJL0 A0A154PJ85 A0A067RFT6 A0A1I8PUQ9 A0A2J7PE96 A0A0P4VY86 A0A023FAP0 A0A1B6GNE2 A0A1B6JV27 A0A182ITT6 A0A1B6DHG3
D6W8L6 C3PPF4 A0A1Y1LWG3 A0A212FD42 A0A182H1V3 B4NHZ7 U4U778 J3JTF2 A0A1W4WMN4 A0A1L8DVW6 B4JGS1 T1E893 A0A2M3Z838 A0A2M4ANV4 A0A2M4C1J1 W5JSZ8 A0A084WSR3 B4QS88 A0A3B0K3P0 Q29BS2 B4GN06 B4PPC3 B3M1X6 B4HGV6 B3P491 Q9VG51 A0A0T6B412 A0A182F8L7 A0A1W4UN59 A0A0K8VXL3 A0A0A1WZP1 A0A034VX97 B4MC73 B4K7G6 B0W379 A0A0A9X9Y5 A0A182QFF7 A0A0M4F4A9 A0A0K8TQE9 A0A1Q3FDW6 A0A182M7V5 A0A182VHS7 A0A182JZF3 A0A182TW52 A0A182VSA8 A0A182XK53 A0A182KX08 Q7QDI2 A0A182R830 A0A182HJT7 A0A182PMX9 U5EI23 T1PC58 A0A151IRM0 A0A195BWJ8 A0A195FQJ7 A0A158NUW6 E1ZWA0 A0A151WT21 A0A2H8TFU9 C4WTD1 A0A0L7R8J5 A0A0L0CGE8 A0A195CRM6 A0A224XV01 A0A069DPS7 R4G544 Q17CA9 E2B7F6 T1D517 A0A1L8EDS5 A0A026W2F6 A0A2A3E517 V9IKW3 A0A0N0BC78 A0A088A6E1 K7IX60 A0A1B0G8S6 A0A1A9XJ17 A0A1A9V744 A0A1B0BXY8 A0A1A9W601 A0A1S3D9U8 A0A336MJL0 A0A154PJ85 A0A067RFT6 A0A1I8PUQ9 A0A2J7PE96 A0A0P4VY86 A0A023FAP0 A0A1B6GNE2 A0A1B6JV27 A0A182ITT6 A0A1B6DHG3
PDB
5F0P
E-value=5.40458e-54,
Score=528
Ontologies
GO
Topology
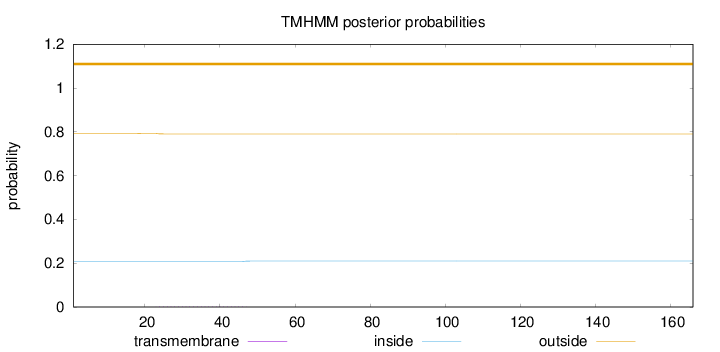
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04658
Exp number, first 60 AAs:
0.04658
Total prob of N-in:
0.20795
outside
1 - 166
Population Genetic Test Statistics
Pi
205.581173
Theta
170.625586
Tajima's D
0.576735
CLR
29.541614
CSRT
0.533473326333683
Interpretation
Uncertain
