Gene
KWMTBOMO13944
Pre Gene Modal
BGIBMGA011292
Annotation
PREDICTED:_uncharacterized_protein_LOC101745580_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.706
Sequence
CDS
ATGTTGATTATCAGTGCCTTAATAACTGGGATGGTGACCGGTCTTCAGAACATTACGACCGAGTCTCCATCGATGATCAGTGACATCACAACATCATTGAATACGAAGGATAAAGTTCCAGAAATGGGGCTGGTTGAACCCACTAAGCTATCTCTACCCACTAGTGACATGGTGAATGTCAGTATTCACGACCAGCCGTCTATGTGGGATTTGTACTTGCACCACTGTTCCAAGTGGAGGCCAATAAATCACATATTTTTCCAAGTGGCGAATATATTCTTTCTTATGTCATTCCTGGCACCTCATACAGACATTGGGCAAATATGGCTTCGGGTGGGCCTGATGATGGGGTGCGCGTTTTCAGGAATGTGGGCATGGAGTGTTGAATGTTACTTGGATGCAGTGCTATGGAACAGTGTATTTATATTGATAAACTTTGTGTATTTCTCTTACCAGTTTTATTTAATGAGACCAATTAAGTTCCACAAGGAGATTGAAGAGGTGTACTTAGCGTTGTTCAAACCACTACGGGTATCAAGGAGACAGTTCCGCCGAGTGCTCATGTGCATGAGAAATGTGAGGCACCTCAAGTGTCACGAACTCTATGCACATGAGAAGGTTACTAAAGTTGACAGCCTTTCGTTGGTGCTTTCGGGCAAGCTTGTGGTGTCACAGAATCAGCGAGCTCTCCATATAGTGTTTCCTCATCATTTCTTGGACTCTCCCGAGTGGTTCGGCGTTTCGACTGACGATTATTTTCAGGTGTCTATAATGGCGATGGAAGAGTCTAGGGTGCTGGTCTGGCATAGAGATAAGCTGAAACTATCAATAATGTCGGACGCTTTCCTGCAAGCGGTCTTCGATCATATCCTCGGACGGGACGTGGTCCACAAATTGATGCAGGTAAGCGAAACGATGGCAGTCAGCAACGGTCACCTCCCTAACAGCTATGAAGAGGGAGAGGACAAGCCGATGCTGGTCGTGAAGAAAGCTGGCGATGGACCTGGAATCACCGCGATACTCAACAGGCAGCTTCAAGGCGAGCATGCGCCGTTACTGCGAGCCACGCCGACGACTCATGAATGCTGA
Protein
MLIISALITGMVTGLQNITTESPSMISDITTSLNTKDKVPEMGLVEPTKLSLPTSDMVNVSIHDQPSMWDLYLHHCSKWRPINHIFFQVANIFFLMSFLAPHTDIGQIWLRVGLMMGCAFSGMWAWSVECYLDAVLWNSVFILINFVYFSYQFYLMRPIKFHKEIEEVYLALFKPLRVSRRQFRRVLMCMRNVRHLKCHELYAHEKVTKVDSLSLVLSGKLVVSQNQRALHIVFPHHFLDSPEWFGVSTDDYFQVSIMAMEESRVLVWHRDKLKLSIMSDAFLQAVFDHILGRDVVHKLMQVSETMAVSNGHLPNSYEEGEDKPMLVVKKAGDGPGITAILNRQLQGEHAPLLRATPTTHEC
Summary
Uniprot
A0A2A4K8U2
L7X0T6
A0A3S2M1B2
A0A194QI18
A0A212F701
A0A1B6LJ07
+ More
A0A1L8D9R7 A0A1L8D9K3 A0A0A9W0V2 A0A0A9W4R3 A0A0A9XNS9 A0A0P4VG13 A0A224XND3 W8AUH3 A0A0A1XC03 A0A034W9V1 A0A026WHE1 A0A1B0CKJ6 A0A0L7QW83 A0A2A3E078 A0A023EW75 A0A1Q3FXU5 A0A1Q3FXP7 A0A1Q3FXL9 A0A1Q3FXT4 A0A1Q3FXN7 F4W724 A0A195F4Y8 A0A195DB53 A0A195AXA3 A0A158NM42 E2B1Q1 A0A1Q3FXN0 A0A154P5S9 A0A1Q3FXP2 A0A1Q3FXX6 A0A1Q3FXQ1 W8BTL2 A0A1Q3FXL4 A0A195C3S6 E2BAC1 A0A088ABM9 A0A0M8ZYP5 A0A151X7X9 A0A1I8NII6 K7IY30 A0A034WAA2 A0A0Q9XR07 A0A0L0BXG7 A0A2C9H899 A0A232FCK4 Q7QEJ2 B4JIP3 B5DNT2 A0A182R0F4 B4M1T7 B4L8Q2 A0A1S4G8D5 A0A182XYD5 A0A1Y9GKG2 A0A0P4WKQ3 A0A1I8NIH9 A0A0M4EU51 A0A1W4VIP6 A0A1I8PUX7 B4NEA5 A0A0R3P6J7 A0A0Q9WCD3 A0A1W4VIG8 A0A3B0J7I0 A0A0R3P1W3 A0A0Q9XQT1 A0A0Q9WMT7 A0A0Q9XUY6 B4I6D7 Q9VRE2 B3N0I8 A0A0P8YEQ2 B4PYM1 A0A1W4VV76 M9PJS1 B3NYA6 G7H830 A0A0R1EC66 A0A1J1HT04 A0A1J1HTH6 A0A1J1HX22 A0A2M3ZKY3 A0A182FIV4 A0A2A3E0U2 A0A2M4AST2 A0A084VIV5 A0A1J1HRS6 A0A2M4BT92 A0A2M4BT22 A0A2M4CGR0
A0A1L8D9R7 A0A1L8D9K3 A0A0A9W0V2 A0A0A9W4R3 A0A0A9XNS9 A0A0P4VG13 A0A224XND3 W8AUH3 A0A0A1XC03 A0A034W9V1 A0A026WHE1 A0A1B0CKJ6 A0A0L7QW83 A0A2A3E078 A0A023EW75 A0A1Q3FXU5 A0A1Q3FXP7 A0A1Q3FXL9 A0A1Q3FXT4 A0A1Q3FXN7 F4W724 A0A195F4Y8 A0A195DB53 A0A195AXA3 A0A158NM42 E2B1Q1 A0A1Q3FXN0 A0A154P5S9 A0A1Q3FXP2 A0A1Q3FXX6 A0A1Q3FXQ1 W8BTL2 A0A1Q3FXL4 A0A195C3S6 E2BAC1 A0A088ABM9 A0A0M8ZYP5 A0A151X7X9 A0A1I8NII6 K7IY30 A0A034WAA2 A0A0Q9XR07 A0A0L0BXG7 A0A2C9H899 A0A232FCK4 Q7QEJ2 B4JIP3 B5DNT2 A0A182R0F4 B4M1T7 B4L8Q2 A0A1S4G8D5 A0A182XYD5 A0A1Y9GKG2 A0A0P4WKQ3 A0A1I8NIH9 A0A0M4EU51 A0A1W4VIP6 A0A1I8PUX7 B4NEA5 A0A0R3P6J7 A0A0Q9WCD3 A0A1W4VIG8 A0A3B0J7I0 A0A0R3P1W3 A0A0Q9XQT1 A0A0Q9WMT7 A0A0Q9XUY6 B4I6D7 Q9VRE2 B3N0I8 A0A0P8YEQ2 B4PYM1 A0A1W4VV76 M9PJS1 B3NYA6 G7H830 A0A0R1EC66 A0A1J1HT04 A0A1J1HTH6 A0A1J1HX22 A0A2M3ZKY3 A0A182FIV4 A0A2A3E0U2 A0A2M4AST2 A0A084VIV5 A0A1J1HRS6 A0A2M4BT92 A0A2M4BT22 A0A2M4CGR0
Pubmed
23674305
26354079
22118469
25401762
26823975
27129103
+ More
24495485 25830018 25348373 24508170 30249741 24945155 21719571 21347285 20798317 25315136 20075255 17994087 26108605 28648823 12364791 15632085 18057021 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588
24495485 25830018 25348373 24508170 30249741 24945155 21719571 21347285 20798317 25315136 20075255 17994087 26108605 28648823 12364791 15632085 18057021 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588
EMBL
NWSH01000057
PCG80100.1
KC469894
AGC92724.1
RSAL01000075
RVE48889.1
+ More
KQ458761 KPJ05157.1 AGBW02009949 OWR49510.1 GEBQ01016295 JAT23682.1 GFDF01011030 JAV03054.1 GFDF01011029 JAV03055.1 GBHO01042135 GBHO01042133 GBRD01000026 GBRD01000025 GDHC01017116 JAG01469.1 JAG01471.1 JAG65795.1 JAQ01513.1 GBHO01042136 JAG01468.1 GBHO01042137 GBHO01024844 GBRD01000024 GBRD01000023 GDHC01009249 JAG01467.1 JAG18760.1 JAG65797.1 JAQ09380.1 GDKW01003546 JAI53049.1 GFTR01005098 JAW11328.1 GAMC01014098 GAMC01014095 JAB92460.1 GBXI01005811 JAD08481.1 GAKP01007870 JAC51082.1 KK107231 QOIP01000009 EZA55081.1 RLU18923.1 AJWK01016334 AJWK01016335 AJWK01016336 KQ414716 KOC62883.1 KZ288499 PBC25165.1 GAPW01000974 JAC12624.1 GFDL01002722 JAV32323.1 GFDL01002717 JAV32328.1 GFDL01002726 JAV32319.1 GFDL01002729 JAV32316.1 GFDL01002708 JAV32337.1 GL887802 EGI70014.1 KQ981805 KYN35650.1 KQ981042 KYN10082.1 KQ976716 KYM76816.1 ADTU01002219 ADTU01002220 GL444953 EFN60407.1 GFDL01002720 JAV32325.1 KQ434823 KZC07227.1 GFDL01002711 JAV32334.1 GFDL01002699 JAV32346.1 GFDL01002707 JAV32338.1 GAMC01014096 JAB92459.1 GFDL01002721 JAV32324.1 KQ978344 KYM94831.1 GL446696 EFN87376.1 KQ435812 KOX72786.1 KQ982431 KYQ56481.1 GAKP01007872 JAC51080.1 CH933815 KRG07554.1 JRES01001179 KNC24753.1 NNAY01000442 OXU28352.1 AAAB01008846 EAA06464.5 CH916370 EDW00490.1 CH379065 EDY73184.1 AXCN02001395 CH940651 EDW65641.1 KRF82325.1 KRF82326.1 EDW08027.1 KRG07557.1 APCN01001015 APCN01001016 GDRN01026227 JAI67691.1 CP012528 ALC48547.1 CH964239 EDW82074.2 KRT07205.1 KRF82323.1 OUUW01000003 SPP78057.1 KRT07204.1 KRG07556.1 KRF82324.1 KRG07555.1 CH480823 EDW56343.1 AE014298 AF247183 AY113513 AAF50859.2 AAG18016.1 AAM29518.1 AAS65416.1 CH902640 EDV38392.2 KPU77391.1 CM000162 EDX03062.1 AGB95591.1 CH954180 EDV47585.2 BT132753 AET07636.1 AGB95590.1 KRK07084.1 CVRI01000020 CRK90682.1 CRK90684.1 CRK90681.1 GGFM01008327 MBW29078.1 PBC25164.1 GGFK01010471 MBW43792.1 ATLV01013410 ATLV01013411 KE524855 KFB37899.1 CRK90683.1 GGFJ01007083 MBW56224.1 GGFJ01007084 MBW56225.1 GGFL01000237 MBW64415.1
KQ458761 KPJ05157.1 AGBW02009949 OWR49510.1 GEBQ01016295 JAT23682.1 GFDF01011030 JAV03054.1 GFDF01011029 JAV03055.1 GBHO01042135 GBHO01042133 GBRD01000026 GBRD01000025 GDHC01017116 JAG01469.1 JAG01471.1 JAG65795.1 JAQ01513.1 GBHO01042136 JAG01468.1 GBHO01042137 GBHO01024844 GBRD01000024 GBRD01000023 GDHC01009249 JAG01467.1 JAG18760.1 JAG65797.1 JAQ09380.1 GDKW01003546 JAI53049.1 GFTR01005098 JAW11328.1 GAMC01014098 GAMC01014095 JAB92460.1 GBXI01005811 JAD08481.1 GAKP01007870 JAC51082.1 KK107231 QOIP01000009 EZA55081.1 RLU18923.1 AJWK01016334 AJWK01016335 AJWK01016336 KQ414716 KOC62883.1 KZ288499 PBC25165.1 GAPW01000974 JAC12624.1 GFDL01002722 JAV32323.1 GFDL01002717 JAV32328.1 GFDL01002726 JAV32319.1 GFDL01002729 JAV32316.1 GFDL01002708 JAV32337.1 GL887802 EGI70014.1 KQ981805 KYN35650.1 KQ981042 KYN10082.1 KQ976716 KYM76816.1 ADTU01002219 ADTU01002220 GL444953 EFN60407.1 GFDL01002720 JAV32325.1 KQ434823 KZC07227.1 GFDL01002711 JAV32334.1 GFDL01002699 JAV32346.1 GFDL01002707 JAV32338.1 GAMC01014096 JAB92459.1 GFDL01002721 JAV32324.1 KQ978344 KYM94831.1 GL446696 EFN87376.1 KQ435812 KOX72786.1 KQ982431 KYQ56481.1 GAKP01007872 JAC51080.1 CH933815 KRG07554.1 JRES01001179 KNC24753.1 NNAY01000442 OXU28352.1 AAAB01008846 EAA06464.5 CH916370 EDW00490.1 CH379065 EDY73184.1 AXCN02001395 CH940651 EDW65641.1 KRF82325.1 KRF82326.1 EDW08027.1 KRG07557.1 APCN01001015 APCN01001016 GDRN01026227 JAI67691.1 CP012528 ALC48547.1 CH964239 EDW82074.2 KRT07205.1 KRF82323.1 OUUW01000003 SPP78057.1 KRT07204.1 KRG07556.1 KRF82324.1 KRG07555.1 CH480823 EDW56343.1 AE014298 AF247183 AY113513 AAF50859.2 AAG18016.1 AAM29518.1 AAS65416.1 CH902640 EDV38392.2 KPU77391.1 CM000162 EDX03062.1 AGB95591.1 CH954180 EDV47585.2 BT132753 AET07636.1 AGB95590.1 KRK07084.1 CVRI01000020 CRK90682.1 CRK90684.1 CRK90681.1 GGFM01008327 MBW29078.1 PBC25164.1 GGFK01010471 MBW43792.1 ATLV01013410 ATLV01013411 KE524855 KFB37899.1 CRK90683.1 GGFJ01007083 MBW56224.1 GGFJ01007084 MBW56225.1 GGFL01000237 MBW64415.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053097
UP000279307
+ More
UP000092461 UP000053825 UP000242457 UP000007755 UP000078541 UP000078492 UP000078540 UP000005205 UP000000311 UP000076502 UP000078542 UP000008237 UP000005203 UP000053105 UP000075809 UP000095301 UP000002358 UP000009192 UP000037069 UP000076407 UP000215335 UP000007062 UP000001070 UP000001819 UP000075886 UP000008792 UP000076408 UP000075840 UP000092553 UP000192221 UP000095300 UP000007798 UP000268350 UP000001292 UP000000803 UP000007801 UP000002282 UP000008711 UP000183832 UP000069272 UP000030765
UP000092461 UP000053825 UP000242457 UP000007755 UP000078541 UP000078492 UP000078540 UP000005205 UP000000311 UP000076502 UP000078542 UP000008237 UP000005203 UP000053105 UP000075809 UP000095301 UP000002358 UP000009192 UP000037069 UP000076407 UP000215335 UP000007062 UP000001070 UP000001819 UP000075886 UP000008792 UP000076408 UP000075840 UP000092553 UP000192221 UP000095300 UP000007798 UP000268350 UP000001292 UP000000803 UP000007801 UP000002282 UP000008711 UP000183832 UP000069272 UP000030765
Pfam
PF04831 Popeye
SUPFAM
SSF51206
SSF51206
Gene 3D
ProteinModelPortal
A0A2A4K8U2
L7X0T6
A0A3S2M1B2
A0A194QI18
A0A212F701
A0A1B6LJ07
+ More
A0A1L8D9R7 A0A1L8D9K3 A0A0A9W0V2 A0A0A9W4R3 A0A0A9XNS9 A0A0P4VG13 A0A224XND3 W8AUH3 A0A0A1XC03 A0A034W9V1 A0A026WHE1 A0A1B0CKJ6 A0A0L7QW83 A0A2A3E078 A0A023EW75 A0A1Q3FXU5 A0A1Q3FXP7 A0A1Q3FXL9 A0A1Q3FXT4 A0A1Q3FXN7 F4W724 A0A195F4Y8 A0A195DB53 A0A195AXA3 A0A158NM42 E2B1Q1 A0A1Q3FXN0 A0A154P5S9 A0A1Q3FXP2 A0A1Q3FXX6 A0A1Q3FXQ1 W8BTL2 A0A1Q3FXL4 A0A195C3S6 E2BAC1 A0A088ABM9 A0A0M8ZYP5 A0A151X7X9 A0A1I8NII6 K7IY30 A0A034WAA2 A0A0Q9XR07 A0A0L0BXG7 A0A2C9H899 A0A232FCK4 Q7QEJ2 B4JIP3 B5DNT2 A0A182R0F4 B4M1T7 B4L8Q2 A0A1S4G8D5 A0A182XYD5 A0A1Y9GKG2 A0A0P4WKQ3 A0A1I8NIH9 A0A0M4EU51 A0A1W4VIP6 A0A1I8PUX7 B4NEA5 A0A0R3P6J7 A0A0Q9WCD3 A0A1W4VIG8 A0A3B0J7I0 A0A0R3P1W3 A0A0Q9XQT1 A0A0Q9WMT7 A0A0Q9XUY6 B4I6D7 Q9VRE2 B3N0I8 A0A0P8YEQ2 B4PYM1 A0A1W4VV76 M9PJS1 B3NYA6 G7H830 A0A0R1EC66 A0A1J1HT04 A0A1J1HTH6 A0A1J1HX22 A0A2M3ZKY3 A0A182FIV4 A0A2A3E0U2 A0A2M4AST2 A0A084VIV5 A0A1J1HRS6 A0A2M4BT92 A0A2M4BT22 A0A2M4CGR0
A0A1L8D9R7 A0A1L8D9K3 A0A0A9W0V2 A0A0A9W4R3 A0A0A9XNS9 A0A0P4VG13 A0A224XND3 W8AUH3 A0A0A1XC03 A0A034W9V1 A0A026WHE1 A0A1B0CKJ6 A0A0L7QW83 A0A2A3E078 A0A023EW75 A0A1Q3FXU5 A0A1Q3FXP7 A0A1Q3FXL9 A0A1Q3FXT4 A0A1Q3FXN7 F4W724 A0A195F4Y8 A0A195DB53 A0A195AXA3 A0A158NM42 E2B1Q1 A0A1Q3FXN0 A0A154P5S9 A0A1Q3FXP2 A0A1Q3FXX6 A0A1Q3FXQ1 W8BTL2 A0A1Q3FXL4 A0A195C3S6 E2BAC1 A0A088ABM9 A0A0M8ZYP5 A0A151X7X9 A0A1I8NII6 K7IY30 A0A034WAA2 A0A0Q9XR07 A0A0L0BXG7 A0A2C9H899 A0A232FCK4 Q7QEJ2 B4JIP3 B5DNT2 A0A182R0F4 B4M1T7 B4L8Q2 A0A1S4G8D5 A0A182XYD5 A0A1Y9GKG2 A0A0P4WKQ3 A0A1I8NIH9 A0A0M4EU51 A0A1W4VIP6 A0A1I8PUX7 B4NEA5 A0A0R3P6J7 A0A0Q9WCD3 A0A1W4VIG8 A0A3B0J7I0 A0A0R3P1W3 A0A0Q9XQT1 A0A0Q9WMT7 A0A0Q9XUY6 B4I6D7 Q9VRE2 B3N0I8 A0A0P8YEQ2 B4PYM1 A0A1W4VV76 M9PJS1 B3NYA6 G7H830 A0A0R1EC66 A0A1J1HT04 A0A1J1HTH6 A0A1J1HX22 A0A2M3ZKY3 A0A182FIV4 A0A2A3E0U2 A0A2M4AST2 A0A084VIV5 A0A1J1HRS6 A0A2M4BT92 A0A2M4BT22 A0A2M4CGR0
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 14,
Likelihood: 0.503325
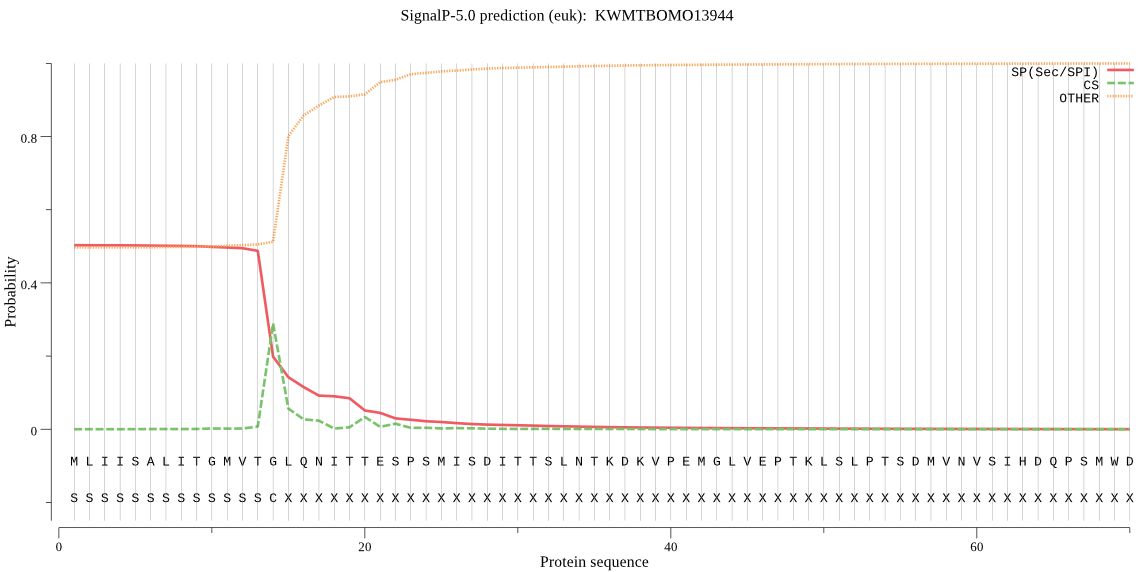
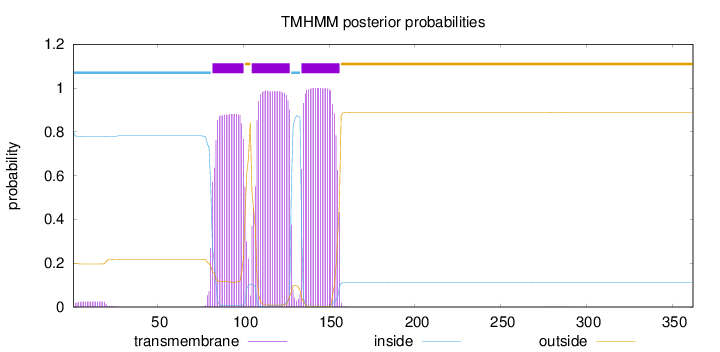
Length:
362
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
60.3561300000001
Exp number, first 60 AAs:
0.46625
Total prob of N-in:
0.80010
inside
1 - 81
TMhelix
82 - 100
outside
101 - 104
TMhelix
105 - 127
inside
128 - 133
TMhelix
134 - 156
outside
157 - 362
Population Genetic Test Statistics
Pi
253.154686
Theta
168.63806
Tajima's D
0.302257
CLR
0.729247
CSRT
0.453777311134443
Interpretation
Uncertain
