Pre Gene Modal
BGIBMGA011314
Annotation
DnaJ-8_[Bombyx_mori]
Full name
DnaJ homolog subfamily C member 25 homolog
Location in the cell
Cytoplasmic Reliability : 2.639
Sequence
CDS
ATGATTACGAGATTTAAATTATGTGTGTTAGTGGGTTTGGCATGGCATTTTATCGAAGTACACAGTCACGATCATCTCCTCGAAGGCATTTACTGTGGGAAACAAAATTGTTACGAAGTTTTGGGTGTAACGCGAGAGGCTACCAAGAATGAGATCGCTAAAAGTTATCGACAACTCGCTAAAAAATTCCATCCGGACTTGCACAGAAAAGCTGAAGATAAAAAAATAGCAGAAGAAAAGTTTAAAGAAATAGCAACGGCCTACGAGATACTACGTGACGATGAAGAGAGGTCTGACTACAATTACATGCTTGACAACCCTCAAGAATATTATGCCCATTACTACCGGTATTACAGACGTCGTATGGCACCTAAAGTCGATGTTAGGATAGTTCTTGCAGTCACTATAACGCTCATATCATTAGTACAGTACTACAGTGCGTGGTCAAAGTATGACACTGCAATAAAATATTTTATGTCAGTTCCAAAATACAGGAACAAGGCATTAGAAATTGCCAAAGAGGAGATAAAAGAGTCACAAGGAGGTAAGAAGAATAGGAAATCAAAAGCAGAACTTAAAAGTGAACAGGATAAAATAATTAGAAGAGTGATTGAAGAAAACATGGACATTAAAGGAGCCTATGCCAAACCTGAAATTGTAGATATTTTGTGGGTTCAATTAATAATTTTGCCCTACACTATAGCATATTACATTTACTGGTATCTGAGATGGATCTGGAAGTACACCATATTAAAACAACCATTTGGCAATGAAGAAAAGTTTTACTTAATTAGGAAATATATGAAAATGGGACCGCACCAGTTTGATGCAATAGAGGAATATGAAAAAAAAGAATACCTTGATGAAGAATTGTGGATAAAAGAAAACTTTAAAATATGGAAAGAGATCAAGGATGAAGAAGCTAAGAAGGCTTTAGCTGATAACAATAAATACAAACAGTACAGAAGGTACATTAAACAACATGGAATCGGGAGGATGACTTTTGATGATTCATGA
Protein
MITRFKLCVLVGLAWHFIEVHSHDHLLEGIYCGKQNCYEVLGVTREATKNEIAKSYRQLAKKFHPDLHRKAEDKKIAEEKFKEIATAYEILRDDEERSDYNYMLDNPQEYYAHYYRYYRRRMAPKVDVRIVLAVTITLISLVQYYSAWSKYDTAIKYFMSVPKYRNKALEIAKEEIKESQGGKKNRKSKAELKSEQDKIIRRVIEENMDIKGAYAKPEIVDILWVQLIILPYTIAYYIYWYLRWIWKYTILKQPFGNEEKFYLIRKYMKMGPHQFDAIEEYEKKEYLDEELWIKENFKIWKEIKDEEAKKALADNNKYKQYRRYIKQHGIGRMTFDDS
Summary
Similarity
Belongs to the DNAJC25 family.
Keywords
Chaperone
Coiled coil
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain DnaJ homolog subfamily C member 25 homolog
Uniprot
H6VTP8
C7AQZ6
A0A0L7LJ03
C3PPE8
A0A194QID7
L7X1R6
+ More
A0A2A4K7P8 A0A194RF46 A0A2H1WUS9 S4P986 D6WD32 I4DLB9 A0A1Y1MTD1 A0A1W4X5V8 A0A1L8DTV3 A0A1L8DTL8 A0A1L8DTM5 A0A1L8DTR2 A0A0L0CBD1 A0A182W5E1 B4MSU4 U5ETT1 A0A1B6CRA4 A0A182MPZ0 A0A2M4BT16 W5JLT6 A0A2M3ZE59 A0A0M5J5S8 A0A182SLG6 A0A2M4ADN0 A0A182PWN7 A0A182Q9D9 A0A182FIT8 A0A182N1J1 D6QX94 A0A182RMT8 A0A1B0D8W5 T1DLG3 A0A336LIK9 A0A182XXX4 A0A1I8NXA1 J3JVP5 A0A212EUD6 A0A182KF95 A0A182G873 A0A182WZ62 A0A182HMN0 A0A182TTY1 Q7Q4X5 A0A182LDD2 A0A182UWH1 B3NTJ5 B4R592 A0A1S4FY71 B4IK88 B4M3K7 Q16K52 Q9VXT2 B4PWP6 W8BH83 A0A1W4V3Q1 B4JLJ2 A0A3B0K5Y5 A0A1D2MUF3 C1BNJ3 Q29I91 B4GWG6 B4L3X0 A0A1Q3G395 B3MZ29 A0A3S1HWI6 A0A2L2Y7S4 A0A226E7B2 V3Z5X4 A0A1B6KEU4 A0A087UWR1 A0A293M484 A0A1B6GPR0 T1J1X2 T1PK45 A0A2G8KJM2 A0A1Z5L1F4 A0A034WK56 A0A1S4EFE1 R7VCT7 A0A1Y3BDE0 A0A0K8VGC9 G3MQH5 V5HZA0 A0A3R7PBD8 A0A224YNK2 A0A131Z5Y0 A0A132AF36 A0A0K2UT64 C3YT47 C1BSF1 A0A023GP47
A0A2A4K7P8 A0A194RF46 A0A2H1WUS9 S4P986 D6WD32 I4DLB9 A0A1Y1MTD1 A0A1W4X5V8 A0A1L8DTV3 A0A1L8DTL8 A0A1L8DTM5 A0A1L8DTR2 A0A0L0CBD1 A0A182W5E1 B4MSU4 U5ETT1 A0A1B6CRA4 A0A182MPZ0 A0A2M4BT16 W5JLT6 A0A2M3ZE59 A0A0M5J5S8 A0A182SLG6 A0A2M4ADN0 A0A182PWN7 A0A182Q9D9 A0A182FIT8 A0A182N1J1 D6QX94 A0A182RMT8 A0A1B0D8W5 T1DLG3 A0A336LIK9 A0A182XXX4 A0A1I8NXA1 J3JVP5 A0A212EUD6 A0A182KF95 A0A182G873 A0A182WZ62 A0A182HMN0 A0A182TTY1 Q7Q4X5 A0A182LDD2 A0A182UWH1 B3NTJ5 B4R592 A0A1S4FY71 B4IK88 B4M3K7 Q16K52 Q9VXT2 B4PWP6 W8BH83 A0A1W4V3Q1 B4JLJ2 A0A3B0K5Y5 A0A1D2MUF3 C1BNJ3 Q29I91 B4GWG6 B4L3X0 A0A1Q3G395 B3MZ29 A0A3S1HWI6 A0A2L2Y7S4 A0A226E7B2 V3Z5X4 A0A1B6KEU4 A0A087UWR1 A0A293M484 A0A1B6GPR0 T1J1X2 T1PK45 A0A2G8KJM2 A0A1Z5L1F4 A0A034WK56 A0A1S4EFE1 R7VCT7 A0A1Y3BDE0 A0A0K8VGC9 G3MQH5 V5HZA0 A0A3R7PBD8 A0A224YNK2 A0A131Z5Y0 A0A132AF36 A0A0K2UT64 C3YT47 C1BSF1 A0A023GP47
Pubmed
19121390
26434795
26227816
26354079
23674305
23622113
+ More
18362917 19820115 22651552 28004739 26108605 17994087 20920257 23761445 24438588 25244985 22516182 23537049 22118469 26483478 12364791 14747013 17210077 20966253 17510324 10731132 12537572 12537569 17550304 24495485 27289101 15632085 26561354 23254933 25315136 29023486 28528879 25348373 22216098 25765539 28797301 26830274 26555130 18563158
18362917 19820115 22651552 28004739 26108605 17994087 20920257 23761445 24438588 25244985 22516182 23537049 22118469 26483478 12364791 14747013 17210077 20966253 17510324 10731132 12537572 12537569 17550304 24495485 27289101 15632085 26561354 23254933 25315136 29023486 28528879 25348373 22216098 25765539 28797301 26830274 26555130 18563158
EMBL
BABH01009572
JN872883
AFC01222.1
FJ592081
ACT34042.1
JTDY01000921
+ More
KOB75427.1 CU467807 CAY54138.1 KQ458761 KPJ05159.1 KC469894 AGC92723.1 NWSH01000057 PCG80099.1 KQ460297 KPJ16207.1 ODYU01011218 SOQ56821.1 GAIX01003844 JAA88716.1 KQ971312 EEZ98325.1 AK402087 BAM18709.1 GEZM01021537 JAV88954.1 GFDF01004274 JAV09810.1 GFDF01004275 JAV09809.1 GFDF01004272 JAV09812.1 GFDF01004273 JAV09811.1 JRES01000749 KNC28774.1 CH963851 EDW75183.1 GANO01001767 JAB58104.1 GEDC01021219 GEDC01001073 JAS16079.1 JAS36225.1 AXCM01000181 GGFJ01007076 MBW56217.1 ADMH02000771 ETN65096.1 GGFM01006051 MBW26802.1 CP012528 ALC48281.1 GGFK01005566 MBW38887.1 AXCN02001449 ATLV01013118 HM013840 KE524840 ADG56643.1 KFB37244.1 AJVK01004157 GAMD01000456 JAB01135.1 UFQT01000014 SSX17686.1 APGK01040556 BT127313 KB740983 KB632410 AEE62275.1 ENN76354.1 ERL95205.1 AGBW02012399 OWR45122.1 JXUM01047497 KQ561521 KXJ78302.1 APCN01004842 AAAB01008963 EAA12149.3 CH954180 EDV46999.1 CM000366 EDX17993.1 CH480852 EDW51480.1 CH940651 EDW65382.1 CH477976 EAT34680.1 AE014298 AY058556 CM000162 EDX01792.1 GAMC01010212 JAB96343.1 CH916370 EDW00445.1 OUUW01000003 SPP78868.1 LJIJ01000509 ODM96717.1 BT076172 ACO10596.1 CH379063 EAL32762.1 CH479194 EDW27050.1 CH933810 EDW07248.1 GFDL01000826 JAV34219.1 CH902632 EDV32873.1 RQTK01000108 RUS87532.1 IAAA01017513 LAA04201.1 LNIX01000006 OXA52837.1 KB203149 ESO86178.1 GEBQ01030011 JAT09966.1 KK122043 KFM81800.1 GFWV01010387 MAA35116.1 GECZ01005351 JAS64418.1 JH431790 KA649049 AFP63678.1 MRZV01000537 PIK48167.1 GFJQ02005952 JAW01018.1 GAKP01002981 JAC55971.1 AMQN01000601 KB293180 ELU16377.1 MUJZ01025605 OTF78941.1 GDHF01014411 JAI37903.1 JO844126 AEO35743.1 GANP01004445 JAB80023.1 QCYY01001061 ROT80829.1 GFPF01007359 MAA18505.1 GEDV01002375 JAP86182.1 JXLN01013593 KPM09469.1 HACA01024091 HACA01024092 CDW41453.1 GG666551 EEN56414.1 BT077530 ACO11954.1 GBBM01000195 JAC35223.1
KOB75427.1 CU467807 CAY54138.1 KQ458761 KPJ05159.1 KC469894 AGC92723.1 NWSH01000057 PCG80099.1 KQ460297 KPJ16207.1 ODYU01011218 SOQ56821.1 GAIX01003844 JAA88716.1 KQ971312 EEZ98325.1 AK402087 BAM18709.1 GEZM01021537 JAV88954.1 GFDF01004274 JAV09810.1 GFDF01004275 JAV09809.1 GFDF01004272 JAV09812.1 GFDF01004273 JAV09811.1 JRES01000749 KNC28774.1 CH963851 EDW75183.1 GANO01001767 JAB58104.1 GEDC01021219 GEDC01001073 JAS16079.1 JAS36225.1 AXCM01000181 GGFJ01007076 MBW56217.1 ADMH02000771 ETN65096.1 GGFM01006051 MBW26802.1 CP012528 ALC48281.1 GGFK01005566 MBW38887.1 AXCN02001449 ATLV01013118 HM013840 KE524840 ADG56643.1 KFB37244.1 AJVK01004157 GAMD01000456 JAB01135.1 UFQT01000014 SSX17686.1 APGK01040556 BT127313 KB740983 KB632410 AEE62275.1 ENN76354.1 ERL95205.1 AGBW02012399 OWR45122.1 JXUM01047497 KQ561521 KXJ78302.1 APCN01004842 AAAB01008963 EAA12149.3 CH954180 EDV46999.1 CM000366 EDX17993.1 CH480852 EDW51480.1 CH940651 EDW65382.1 CH477976 EAT34680.1 AE014298 AY058556 CM000162 EDX01792.1 GAMC01010212 JAB96343.1 CH916370 EDW00445.1 OUUW01000003 SPP78868.1 LJIJ01000509 ODM96717.1 BT076172 ACO10596.1 CH379063 EAL32762.1 CH479194 EDW27050.1 CH933810 EDW07248.1 GFDL01000826 JAV34219.1 CH902632 EDV32873.1 RQTK01000108 RUS87532.1 IAAA01017513 LAA04201.1 LNIX01000006 OXA52837.1 KB203149 ESO86178.1 GEBQ01030011 JAT09966.1 KK122043 KFM81800.1 GFWV01010387 MAA35116.1 GECZ01005351 JAS64418.1 JH431790 KA649049 AFP63678.1 MRZV01000537 PIK48167.1 GFJQ02005952 JAW01018.1 GAKP01002981 JAC55971.1 AMQN01000601 KB293180 ELU16377.1 MUJZ01025605 OTF78941.1 GDHF01014411 JAI37903.1 JO844126 AEO35743.1 GANP01004445 JAB80023.1 QCYY01001061 ROT80829.1 GFPF01007359 MAA18505.1 GEDV01002375 JAP86182.1 JXLN01013593 KPM09469.1 HACA01024091 HACA01024092 CDW41453.1 GG666551 EEN56414.1 BT077530 ACO11954.1 GBBM01000195 JAC35223.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000218220
UP000053240
UP000007266
+ More
UP000192223 UP000037069 UP000075920 UP000007798 UP000075883 UP000000673 UP000092553 UP000075901 UP000075885 UP000075886 UP000069272 UP000075884 UP000030765 UP000075900 UP000092462 UP000076408 UP000095300 UP000019118 UP000030742 UP000007151 UP000075881 UP000069940 UP000249989 UP000076407 UP000075840 UP000075902 UP000007062 UP000075882 UP000075903 UP000008711 UP000000304 UP000001292 UP000008792 UP000008820 UP000000803 UP000002282 UP000192221 UP000001070 UP000268350 UP000094527 UP000001819 UP000008744 UP000009192 UP000007801 UP000271974 UP000198287 UP000030746 UP000054359 UP000095301 UP000230750 UP000079169 UP000014760 UP000283509 UP000001554
UP000192223 UP000037069 UP000075920 UP000007798 UP000075883 UP000000673 UP000092553 UP000075901 UP000075885 UP000075886 UP000069272 UP000075884 UP000030765 UP000075900 UP000092462 UP000076408 UP000095300 UP000019118 UP000030742 UP000007151 UP000075881 UP000069940 UP000249989 UP000076407 UP000075840 UP000075902 UP000007062 UP000075882 UP000075903 UP000008711 UP000000304 UP000001292 UP000008792 UP000008820 UP000000803 UP000002282 UP000192221 UP000001070 UP000268350 UP000094527 UP000001819 UP000008744 UP000009192 UP000007801 UP000271974 UP000198287 UP000030746 UP000054359 UP000095301 UP000230750 UP000079169 UP000014760 UP000283509 UP000001554
Interpro
Gene 3D
CDD
ProteinModelPortal
H6VTP8
C7AQZ6
A0A0L7LJ03
C3PPE8
A0A194QID7
L7X1R6
+ More
A0A2A4K7P8 A0A194RF46 A0A2H1WUS9 S4P986 D6WD32 I4DLB9 A0A1Y1MTD1 A0A1W4X5V8 A0A1L8DTV3 A0A1L8DTL8 A0A1L8DTM5 A0A1L8DTR2 A0A0L0CBD1 A0A182W5E1 B4MSU4 U5ETT1 A0A1B6CRA4 A0A182MPZ0 A0A2M4BT16 W5JLT6 A0A2M3ZE59 A0A0M5J5S8 A0A182SLG6 A0A2M4ADN0 A0A182PWN7 A0A182Q9D9 A0A182FIT8 A0A182N1J1 D6QX94 A0A182RMT8 A0A1B0D8W5 T1DLG3 A0A336LIK9 A0A182XXX4 A0A1I8NXA1 J3JVP5 A0A212EUD6 A0A182KF95 A0A182G873 A0A182WZ62 A0A182HMN0 A0A182TTY1 Q7Q4X5 A0A182LDD2 A0A182UWH1 B3NTJ5 B4R592 A0A1S4FY71 B4IK88 B4M3K7 Q16K52 Q9VXT2 B4PWP6 W8BH83 A0A1W4V3Q1 B4JLJ2 A0A3B0K5Y5 A0A1D2MUF3 C1BNJ3 Q29I91 B4GWG6 B4L3X0 A0A1Q3G395 B3MZ29 A0A3S1HWI6 A0A2L2Y7S4 A0A226E7B2 V3Z5X4 A0A1B6KEU4 A0A087UWR1 A0A293M484 A0A1B6GPR0 T1J1X2 T1PK45 A0A2G8KJM2 A0A1Z5L1F4 A0A034WK56 A0A1S4EFE1 R7VCT7 A0A1Y3BDE0 A0A0K8VGC9 G3MQH5 V5HZA0 A0A3R7PBD8 A0A224YNK2 A0A131Z5Y0 A0A132AF36 A0A0K2UT64 C3YT47 C1BSF1 A0A023GP47
A0A2A4K7P8 A0A194RF46 A0A2H1WUS9 S4P986 D6WD32 I4DLB9 A0A1Y1MTD1 A0A1W4X5V8 A0A1L8DTV3 A0A1L8DTL8 A0A1L8DTM5 A0A1L8DTR2 A0A0L0CBD1 A0A182W5E1 B4MSU4 U5ETT1 A0A1B6CRA4 A0A182MPZ0 A0A2M4BT16 W5JLT6 A0A2M3ZE59 A0A0M5J5S8 A0A182SLG6 A0A2M4ADN0 A0A182PWN7 A0A182Q9D9 A0A182FIT8 A0A182N1J1 D6QX94 A0A182RMT8 A0A1B0D8W5 T1DLG3 A0A336LIK9 A0A182XXX4 A0A1I8NXA1 J3JVP5 A0A212EUD6 A0A182KF95 A0A182G873 A0A182WZ62 A0A182HMN0 A0A182TTY1 Q7Q4X5 A0A182LDD2 A0A182UWH1 B3NTJ5 B4R592 A0A1S4FY71 B4IK88 B4M3K7 Q16K52 Q9VXT2 B4PWP6 W8BH83 A0A1W4V3Q1 B4JLJ2 A0A3B0K5Y5 A0A1D2MUF3 C1BNJ3 Q29I91 B4GWG6 B4L3X0 A0A1Q3G395 B3MZ29 A0A3S1HWI6 A0A2L2Y7S4 A0A226E7B2 V3Z5X4 A0A1B6KEU4 A0A087UWR1 A0A293M484 A0A1B6GPR0 T1J1X2 T1PK45 A0A2G8KJM2 A0A1Z5L1F4 A0A034WK56 A0A1S4EFE1 R7VCT7 A0A1Y3BDE0 A0A0K8VGC9 G3MQH5 V5HZA0 A0A3R7PBD8 A0A224YNK2 A0A131Z5Y0 A0A132AF36 A0A0K2UT64 C3YT47 C1BSF1 A0A023GP47
PDB
2QSA
E-value=6.89597e-12,
Score=170
Ontologies
Topology
Subcellular location
Membrane
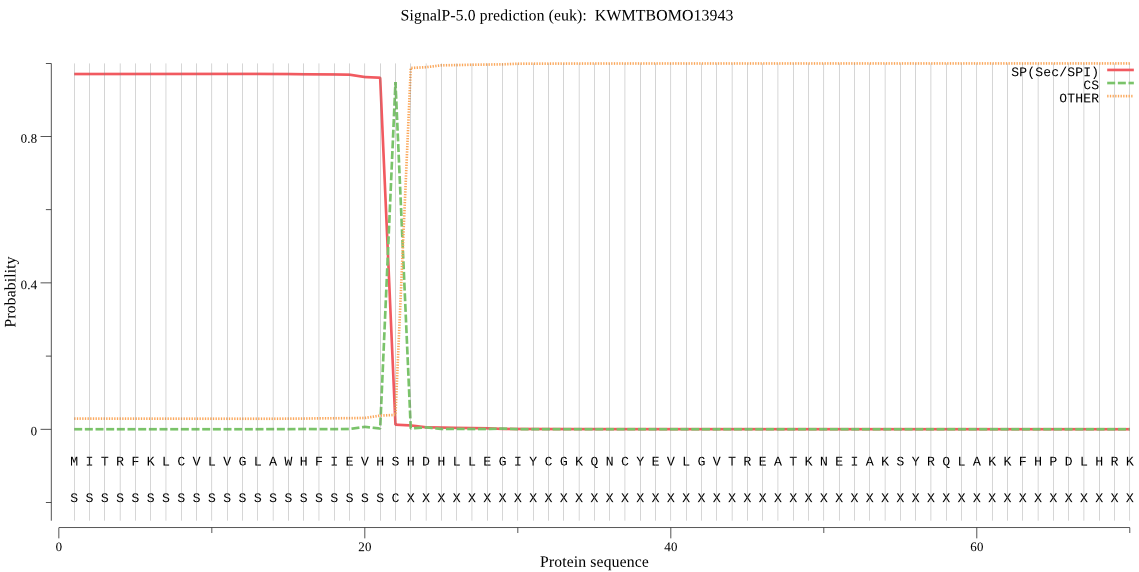
SignalP
Position: 1 - 22,
Likelihood: 0.970601
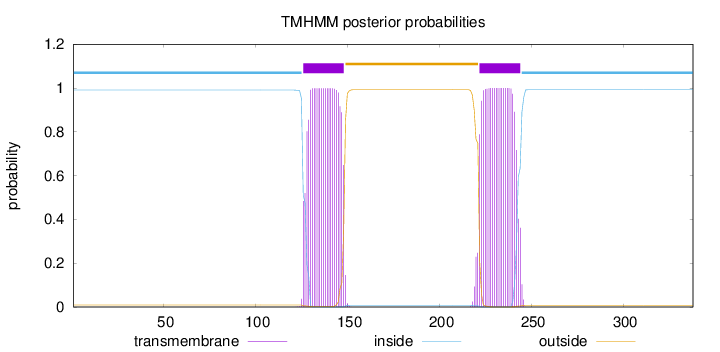
Length:
338
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.05015
Exp number, first 60 AAs:
0.00206
Total prob of N-in:
0.99082
inside
1 - 125
TMhelix
126 - 148
outside
149 - 221
TMhelix
222 - 244
inside
245 - 338
Population Genetic Test Statistics
Pi
250.831809
Theta
234.228321
Tajima's D
0.204877
CLR
13.43797
CSRT
0.424778761061947
Interpretation
Uncertain
