Gene
KWMTBOMO13941
Annotation
PREDICTED:_max_dimerization_protein_4-like_isoform_X1_[Bombyx_mori]
Full name
Max dimerization protein 1
Alternative Name
Protein MAD
Location in the cell
Mitochondrial Reliability : 1.307 Nuclear Reliability : 1.439
Sequence
CDS
ATGAGCATTGCCGATTTAATCCAAGCCGCCGAATTCATCGAACGCCGTGACCGAGTTTTGTTCATTGCAGACGCGGAACACGGTTACGCCTCGGCGTTACCTATATACGAAGATCCCAAGCCGGGGGGGAGACGTCCGAAGACGAAAAAATCGCAGGGAAGCAGGACTACACATAACGAATTAGAAAAGAACAGGGTTTTCCGAGTCATAGACGTAATTGAAACTGCTTTAGCGACGACAGTGTCGTTCGCTACCACGAAAACTGTAAAGTATTGA
Protein
MSIADLIQAAEFIERRDRVLFIADAEHGYASALPIYEDPKPGGRRPKTKKSQGSRTTHNELEKNRVFRVIDVIETALATTVSFATTKTVKY
Summary
Subunit
Efficient DNA binding requires dimerization with another bHLH protein. Binds DNA as a heterodimer with MAX. Interacts with RNF17.
Keywords
Complete proteome
DNA-binding
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Ubl conjugation
Feature
chain Max dimerization protein 1
Uniprot
A0A1E1VXN4
A0A2A4K741
A0A212EUE8
C3PPE6
L7X1I6
A0A2H1WUU0
+ More
S4PQT4 E9MYK4 C9E266 E9MYK5 E9MYK7 E9MYK3 A0A2A4K870 E9MYK6 K7INS8 A0A2A3EEX5 L7X3Y5 A0A0L7KR30 V9I9B8 Q14VH3 A0A0C9QHP1 A0A3L8DRR2 A0A088A6X1 A0A158NZG8 A0A2J7PED3 A0A067RH84 A0A2P6KHR1 J9JXQ7 A0A1Y1NFL9 A0A195BPD9 A0A1B6DNF4 A0A2H8TQB3 A0A1B6J9H4 A0A1B6MHE4 A0A1B6GXU6 A0A0A9Z1J8 A0A224XWA8 A0A069DXR5 R4G3H0 A0A1W4X5T8 A0A0B6ZND2 A0A1S3JTA8 A0A293N7R2 A0A2P8Z0J9 A0A023FHU8 A0A2R5L5F2 A0A131Z355 A0A131XRQ6 T1KE75 A0A336MH03 V3Z6W0 A0A2S2R7Y6 A0A139WMI1 R7TJD7 A0A2T7PAG0 A0A2C9LZZ3 A0A3S0ZZ44 A0A0C9RC62 V5HRL2 A0A1B0GJL3 A0A1L8DI79 A0A1J1HHR1 A0A1J1HI17 A0A0P4WIU9 A0A210QL69 A0A3B1J7M0 A0A0N4SW68 K1RGW6 A0A1V4KR28 E9FXP6 A0A164VP50 Q4SJP9 A0A0P6ISW9 F7DZC4 A0A3Q3W5S2 A0A3Q3IW04 H3CUA9 B5G2N4 A0A226PN99 A0A1A8QHP8 A0A226N171 A0A3Q1GGR5 Q8K1Z8 A0A3Q2XGF7 Q4FK19 P50538 A0A0S7H5I3 A4IH47 A0A3Q4HQ93 A0A1L8HKS2 A0A1J1HJ02 I3KMT1
S4PQT4 E9MYK4 C9E266 E9MYK5 E9MYK7 E9MYK3 A0A2A4K870 E9MYK6 K7INS8 A0A2A3EEX5 L7X3Y5 A0A0L7KR30 V9I9B8 Q14VH3 A0A0C9QHP1 A0A3L8DRR2 A0A088A6X1 A0A158NZG8 A0A2J7PED3 A0A067RH84 A0A2P6KHR1 J9JXQ7 A0A1Y1NFL9 A0A195BPD9 A0A1B6DNF4 A0A2H8TQB3 A0A1B6J9H4 A0A1B6MHE4 A0A1B6GXU6 A0A0A9Z1J8 A0A224XWA8 A0A069DXR5 R4G3H0 A0A1W4X5T8 A0A0B6ZND2 A0A1S3JTA8 A0A293N7R2 A0A2P8Z0J9 A0A023FHU8 A0A2R5L5F2 A0A131Z355 A0A131XRQ6 T1KE75 A0A336MH03 V3Z6W0 A0A2S2R7Y6 A0A139WMI1 R7TJD7 A0A2T7PAG0 A0A2C9LZZ3 A0A3S0ZZ44 A0A0C9RC62 V5HRL2 A0A1B0GJL3 A0A1L8DI79 A0A1J1HHR1 A0A1J1HI17 A0A0P4WIU9 A0A210QL69 A0A3B1J7M0 A0A0N4SW68 K1RGW6 A0A1V4KR28 E9FXP6 A0A164VP50 Q4SJP9 A0A0P6ISW9 F7DZC4 A0A3Q3W5S2 A0A3Q3IW04 H3CUA9 B5G2N4 A0A226PN99 A0A1A8QHP8 A0A226N171 A0A3Q1GGR5 Q8K1Z8 A0A3Q2XGF7 Q4FK19 P50538 A0A0S7H5I3 A4IH47 A0A3Q4HQ93 A0A1L8HKS2 A0A1J1HJ02 I3KMT1
Pubmed
22118469
23674305
23622113
21114846
20075255
26227816
+ More
17506851 30249741 21347285 24845553 28004739 25401762 26823975 26334808 29403074 26830274 28049606 23254933 18362917 19820115 15562597 25765539 28812685 25329095 12040188 19468303 22992520 21292972 15496914 20431018 17018643 24621616 10349636 11042159 11076861 11217851 12466851 15489334 16141073 7667316 9519870 7896882 8521822 8617250 10597267 25186727 27762356
17506851 30249741 21347285 24845553 28004739 25401762 26823975 26334808 29403074 26830274 28049606 23254933 18362917 19820115 15562597 25765539 28812685 25329095 12040188 19468303 22992520 21292972 15496914 20431018 17018643 24621616 10349636 11042159 11076861 11217851 12466851 15489334 16141073 7667316 9519870 7896882 8521822 8617250 10597267 25186727 27762356
EMBL
GDQN01011574
JAT79480.1
NWSH01000057
PCG80095.1
AGBW02012399
OWR45120.1
+ More
CU467807 CAY54136.1 KC469894 AGC92721.1 ODYU01011218 SOQ56823.1 GAIX01001405 JAA91155.1 HQ328820 ADT64098.1 GQ452003 ACV83776.1 HQ328821 ADT64099.1 HQ328823 ADT64101.1 HQ328819 ADT64097.1 PCG80094.1 HQ328822 ADT64100.1 KZ288266 PBC30287.1 AGC92720.1 JTDY01007080 KOB65469.1 JR036581 AEY57242.1 DQ666693 ABG29167.1 GBYB01014172 GBYB01014173 JAG83939.1 JAG83940.1 QOIP01000005 RLU22992.1 ADTU01004596 ADTU01004597 ADTU01004598 ADTU01004599 ADTU01004600 NEVH01026101 PNF14689.1 KK852508 KDR22408.1 MWRG01011629 PRD25859.1 ABLF02038824 ABLF02038830 ABLF02038831 ABLF02038835 ABLF02038836 ABLF02038838 GEZM01003720 GEZM01003719 JAV96611.1 KQ976433 KYM87319.1 GEDC01010082 JAS27216.1 GFXV01003643 MBW15448.1 GECU01011849 JAS95857.1 GEBQ01027718 GEBQ01004658 JAT12259.1 JAT35319.1 GECZ01005454 GECZ01002552 JAS64315.1 JAS67217.1 GBHO01033341 GBHO01005254 GBHO01005253 GBHO01005250 GBHO01005248 GBHO01005247 GBRD01009207 GDHC01017829 GDHC01014884 JAG10263.1 JAG38350.1 JAG38351.1 JAG38354.1 JAG38356.1 JAG38357.1 JAG56614.1 JAQ00800.1 JAQ03745.1 GFTR01003656 JAW12770.1 GBGD01002740 JAC86149.1 ACPB03019618 GAHY01001686 JAA75824.1 HACG01023299 CEK70164.1 GFWV01022943 MAA47670.1 PYGN01000254 PSN50022.1 GBBK01003793 JAC20689.1 GGLE01000577 MBY04703.1 GEDV01003387 JAP85170.1 GEFH01000205 JAP68376.1 CAEY01002034 UFQT01001253 SSX29662.1 KB203049 ESO86553.1 GGMS01016885 MBY86088.1 KQ971312 KYB29051.1 AMQN01002628 KB309694 ELT93617.1 PZQS01000005 PVD30410.1 RQTK01000037 RUS90233.1 GBYB01014174 JAG83941.1 GANP01005921 JAB78547.1 AJWK01021928 AJWK01021929 AJWK01021930 AJWK01021931 AJWK01021932 GFDF01007906 JAV06178.1 CVRI01000004 CRK87541.1 CRK87539.1 GDRN01086386 JAI61188.1 NEDP02003088 OWF49474.1 AC158662 CH466523 EDK99189.1 JH816523 EKC40670.1 LSYS01002146 OPJ86863.1 GL732526 EFX88246.1 LRGB01001361 KZS12515.1 CAAE01014573 CAF99133.1 GDIQ01000541 JAN94196.1 AAMC01129610 AAMC01129611 DQ216229 ACH45545.1 AWGT02000041 OXB80982.1 HAEH01011594 SBR92634.1 MCFN01000278 OXB61333.1 BC034860 AK151014 AK151114 AK151305 AK151882 AK153303 AAH34860.1 BAE30034.1 CT010233 CAJ18441.1 L38926 U20614 X83106 GBYX01445448 JAO35984.1 BC135370 AAI35371.1 CM004467 OCT96666.1 CRK87538.1 AERX01014075
CU467807 CAY54136.1 KC469894 AGC92721.1 ODYU01011218 SOQ56823.1 GAIX01001405 JAA91155.1 HQ328820 ADT64098.1 GQ452003 ACV83776.1 HQ328821 ADT64099.1 HQ328823 ADT64101.1 HQ328819 ADT64097.1 PCG80094.1 HQ328822 ADT64100.1 KZ288266 PBC30287.1 AGC92720.1 JTDY01007080 KOB65469.1 JR036581 AEY57242.1 DQ666693 ABG29167.1 GBYB01014172 GBYB01014173 JAG83939.1 JAG83940.1 QOIP01000005 RLU22992.1 ADTU01004596 ADTU01004597 ADTU01004598 ADTU01004599 ADTU01004600 NEVH01026101 PNF14689.1 KK852508 KDR22408.1 MWRG01011629 PRD25859.1 ABLF02038824 ABLF02038830 ABLF02038831 ABLF02038835 ABLF02038836 ABLF02038838 GEZM01003720 GEZM01003719 JAV96611.1 KQ976433 KYM87319.1 GEDC01010082 JAS27216.1 GFXV01003643 MBW15448.1 GECU01011849 JAS95857.1 GEBQ01027718 GEBQ01004658 JAT12259.1 JAT35319.1 GECZ01005454 GECZ01002552 JAS64315.1 JAS67217.1 GBHO01033341 GBHO01005254 GBHO01005253 GBHO01005250 GBHO01005248 GBHO01005247 GBRD01009207 GDHC01017829 GDHC01014884 JAG10263.1 JAG38350.1 JAG38351.1 JAG38354.1 JAG38356.1 JAG38357.1 JAG56614.1 JAQ00800.1 JAQ03745.1 GFTR01003656 JAW12770.1 GBGD01002740 JAC86149.1 ACPB03019618 GAHY01001686 JAA75824.1 HACG01023299 CEK70164.1 GFWV01022943 MAA47670.1 PYGN01000254 PSN50022.1 GBBK01003793 JAC20689.1 GGLE01000577 MBY04703.1 GEDV01003387 JAP85170.1 GEFH01000205 JAP68376.1 CAEY01002034 UFQT01001253 SSX29662.1 KB203049 ESO86553.1 GGMS01016885 MBY86088.1 KQ971312 KYB29051.1 AMQN01002628 KB309694 ELT93617.1 PZQS01000005 PVD30410.1 RQTK01000037 RUS90233.1 GBYB01014174 JAG83941.1 GANP01005921 JAB78547.1 AJWK01021928 AJWK01021929 AJWK01021930 AJWK01021931 AJWK01021932 GFDF01007906 JAV06178.1 CVRI01000004 CRK87541.1 CRK87539.1 GDRN01086386 JAI61188.1 NEDP02003088 OWF49474.1 AC158662 CH466523 EDK99189.1 JH816523 EKC40670.1 LSYS01002146 OPJ86863.1 GL732526 EFX88246.1 LRGB01001361 KZS12515.1 CAAE01014573 CAF99133.1 GDIQ01000541 JAN94196.1 AAMC01129610 AAMC01129611 DQ216229 ACH45545.1 AWGT02000041 OXB80982.1 HAEH01011594 SBR92634.1 MCFN01000278 OXB61333.1 BC034860 AK151014 AK151114 AK151305 AK151882 AK153303 AAH34860.1 BAE30034.1 CT010233 CAJ18441.1 L38926 U20614 X83106 GBYX01445448 JAO35984.1 BC135370 AAI35371.1 CM004467 OCT96666.1 CRK87538.1 AERX01014075
Proteomes
UP000218220
UP000007151
UP000002358
UP000242457
UP000037510
UP000279307
+ More
UP000005203 UP000005205 UP000235965 UP000027135 UP000007819 UP000078540 UP000015103 UP000192223 UP000085678 UP000245037 UP000015104 UP000030746 UP000007266 UP000014760 UP000245119 UP000076420 UP000271974 UP000092461 UP000183832 UP000242188 UP000018467 UP000000589 UP000005408 UP000190648 UP000000305 UP000076858 UP000008143 UP000261620 UP000261600 UP000007303 UP000198419 UP000198323 UP000257200 UP000264820 UP000261580 UP000186698 UP000005207
UP000005203 UP000005205 UP000235965 UP000027135 UP000007819 UP000078540 UP000015103 UP000192223 UP000085678 UP000245037 UP000015104 UP000030746 UP000007266 UP000014760 UP000245119 UP000076420 UP000271974 UP000092461 UP000183832 UP000242188 UP000018467 UP000000589 UP000005408 UP000190648 UP000000305 UP000076858 UP000008143 UP000261620 UP000261600 UP000007303 UP000198419 UP000198323 UP000257200 UP000264820 UP000261580 UP000186698 UP000005207
PRIDE
Interpro
SUPFAM
SSF47459
SSF47459
Gene 3D
CDD
ProteinModelPortal
A0A1E1VXN4
A0A2A4K741
A0A212EUE8
C3PPE6
L7X1I6
A0A2H1WUU0
+ More
S4PQT4 E9MYK4 C9E266 E9MYK5 E9MYK7 E9MYK3 A0A2A4K870 E9MYK6 K7INS8 A0A2A3EEX5 L7X3Y5 A0A0L7KR30 V9I9B8 Q14VH3 A0A0C9QHP1 A0A3L8DRR2 A0A088A6X1 A0A158NZG8 A0A2J7PED3 A0A067RH84 A0A2P6KHR1 J9JXQ7 A0A1Y1NFL9 A0A195BPD9 A0A1B6DNF4 A0A2H8TQB3 A0A1B6J9H4 A0A1B6MHE4 A0A1B6GXU6 A0A0A9Z1J8 A0A224XWA8 A0A069DXR5 R4G3H0 A0A1W4X5T8 A0A0B6ZND2 A0A1S3JTA8 A0A293N7R2 A0A2P8Z0J9 A0A023FHU8 A0A2R5L5F2 A0A131Z355 A0A131XRQ6 T1KE75 A0A336MH03 V3Z6W0 A0A2S2R7Y6 A0A139WMI1 R7TJD7 A0A2T7PAG0 A0A2C9LZZ3 A0A3S0ZZ44 A0A0C9RC62 V5HRL2 A0A1B0GJL3 A0A1L8DI79 A0A1J1HHR1 A0A1J1HI17 A0A0P4WIU9 A0A210QL69 A0A3B1J7M0 A0A0N4SW68 K1RGW6 A0A1V4KR28 E9FXP6 A0A164VP50 Q4SJP9 A0A0P6ISW9 F7DZC4 A0A3Q3W5S2 A0A3Q3IW04 H3CUA9 B5G2N4 A0A226PN99 A0A1A8QHP8 A0A226N171 A0A3Q1GGR5 Q8K1Z8 A0A3Q2XGF7 Q4FK19 P50538 A0A0S7H5I3 A4IH47 A0A3Q4HQ93 A0A1L8HKS2 A0A1J1HJ02 I3KMT1
S4PQT4 E9MYK4 C9E266 E9MYK5 E9MYK7 E9MYK3 A0A2A4K870 E9MYK6 K7INS8 A0A2A3EEX5 L7X3Y5 A0A0L7KR30 V9I9B8 Q14VH3 A0A0C9QHP1 A0A3L8DRR2 A0A088A6X1 A0A158NZG8 A0A2J7PED3 A0A067RH84 A0A2P6KHR1 J9JXQ7 A0A1Y1NFL9 A0A195BPD9 A0A1B6DNF4 A0A2H8TQB3 A0A1B6J9H4 A0A1B6MHE4 A0A1B6GXU6 A0A0A9Z1J8 A0A224XWA8 A0A069DXR5 R4G3H0 A0A1W4X5T8 A0A0B6ZND2 A0A1S3JTA8 A0A293N7R2 A0A2P8Z0J9 A0A023FHU8 A0A2R5L5F2 A0A131Z355 A0A131XRQ6 T1KE75 A0A336MH03 V3Z6W0 A0A2S2R7Y6 A0A139WMI1 R7TJD7 A0A2T7PAG0 A0A2C9LZZ3 A0A3S0ZZ44 A0A0C9RC62 V5HRL2 A0A1B0GJL3 A0A1L8DI79 A0A1J1HHR1 A0A1J1HI17 A0A0P4WIU9 A0A210QL69 A0A3B1J7M0 A0A0N4SW68 K1RGW6 A0A1V4KR28 E9FXP6 A0A164VP50 Q4SJP9 A0A0P6ISW9 F7DZC4 A0A3Q3W5S2 A0A3Q3IW04 H3CUA9 B5G2N4 A0A226PN99 A0A1A8QHP8 A0A226N171 A0A3Q1GGR5 Q8K1Z8 A0A3Q2XGF7 Q4FK19 P50538 A0A0S7H5I3 A4IH47 A0A3Q4HQ93 A0A1L8HKS2 A0A1J1HJ02 I3KMT1
Ontologies
GO
PANTHER
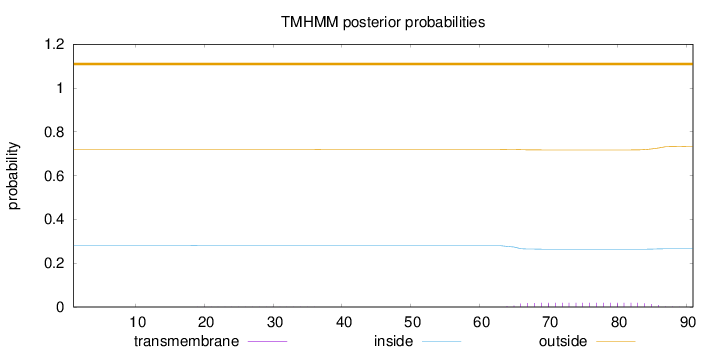
Topology
Subcellular location
Nucleus
Length:
91
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.39506
Exp number, first 60 AAs:
0.00306
Total prob of N-in:
0.28043
outside
1 - 91
Population Genetic Test Statistics
Pi
221.545826
Theta
190.329144
Tajima's D
0.498253
CLR
0
CSRT
0.513224338783061
Interpretation
Uncertain
