Gene
KWMTBOMO13926 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011294
Annotation
PREDICTED:_sideroflexin-2_[Papilio_xuthus]
Full name
Sidoreflexin
+ More
Sideroflexin-2
Sideroflexin-2
Location in the cell
PlasmaMembrane Reliability : 3.005
Sequence
CDS
ATGCCTGAAGAAAAGAGAATAGACATCACCCAACCACTGTGGGACCAGAGCACATTTGTGGGAAGGTTCCGACATTTTGCCTTCATTTCCAATCCACTGCTGTCAATGGCTTCTGAGAAAGAACTGTATGAAGCTAAAGAATTATATCTAAACTACAAAGCAGGTCGTGAGCAGCCAGGGACAAAATTAGAGCAAGTAATTAAGGCGAAACAGCTTTATGAATCAGCATTCCATCCAGACAGCGGGGAGCTGCAGAATGTTTTCGGTAGAATGTCATTCCAAATGCCAGGCGGTTGTCTTATTACCGGTGCTATGTTACAGTGGTACAGAACAGCAACAGCCGTAGTCTTCTGGCAATGGGTGAACCAGTCTTTCAATGCTCTAGTAAACTACACAAATAGGAATGCCAATTCTCCCCTAACCACCAAACAGATGGGAGTGGCATACGTGTCTGCGACATCAGCTGCCATGACCACGGCCCTGATCTTCAAGTTCGGTGTTCAGAAAAGAGCGAAGAATCCTATTTTGGCTCGCTTTGTGCCGTTCGTCGCGGTGGCGGCCGCTAACTGGGTAAACATCCCGCTAATGAGGCAGAACGAAATCATTTTAGGTCTGGACGTCTCTGACGAGAGCGGGAGGGTCATCGGGAAGTCTCAGATCGCACCCGTCAAGGGGATCTCGCAAGTCGTCACGTCGCGGATCATAATGTGCGCACCGGGAATGCTGCTCCTGCCGGTGATAATGGAGAAACTGGAGCCGAAACCCTGGATGCAGAGGATCCGGTGGGCGCACATCGGCATACAGACCGCCATTGTGGGCATGTTCTTGACGTTCATGGTGCCGACTGCGTGCGCGATATTCCCGCAGAAATGCAAACTCTCCATTGACACAATAAAACGTTTAGAGAAAGATAATTATGAAGAAATATTGAGAAACACAGGCGGTAGTCCACCGCCATACGTCTACTTCAACAAGGGCTTGTAG
Protein
MPEEKRIDITQPLWDQSTFVGRFRHFAFISNPLLSMASEKELYEAKELYLNYKAGREQPGTKLEQVIKAKQLYESAFHPDSGELQNVFGRMSFQMPGGCLITGAMLQWYRTATAVVFWQWVNQSFNALVNYTNRNANSPLTTKQMGVAYVSATSAAMTTALIFKFGVQKRAKNPILARFVPFVAVAAANWVNIPLMRQNEIILGLDVSDESGRVIGKSQIAPVKGISQVVTSRIIMCAPGMLLLPVIMEKLEPKPWMQRIRWAHIGIQTAIVGMFLTFMVPTACAIFPQKCKLSIDTIKRLEKDNYEEILRNTGGSPPPYVYFNKGL
Summary
Description
Mitochondrial amino-acid transporter that mediates transport of serine into mitochondria.
Similarity
Belongs to the sideroflexin family.
Keywords
Amino-acid transport
Complete proteome
Membrane
Mitochondrion
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Sideroflexin-2
Uniprot
A0A2A4J177
A0A212EJE5
A0A0N1IE59
A0A194QEA8
A0A1E1WB48
L7X3Y2
+ More
A0A1L8DZU1 A0A1L8DZU2 A0A1L8E039 A0A1B0CIK1 U5EUP8 A0A1W4X7R8 D7EIS6 A0A0N0BFF2 A0A1Y1M0T6 A0A067QH17 A0A088A4I8 A0A1J1HN73 J3JXU8 A0A2A3EEW6 A0A151JMQ4 A0A154PLW9 A0A151XDE3 A0A158NH46 A0A2J7QSX6 N6U2G2 K7INS3 A0A232FEJ9 A0A195D1C1 E9IMV8 A0A182V663 A0A182TWA3 A0A182KQE2 A0A182WWY6 Q7QIX5 A0A182I2I5 A0A182JND0 A0A182P0N0 A0A1A9XK37 R4WDH4 A0A182RP57 A0A2C9GW75 A0A182NK62 A0A182YEK7 A0A182SW02 A0A1I8PE01 A0A182WLI0 E9HAV9 A0A026X2I9 A0A182MNN6 A0A0T6B1A2 A0A1B6CGB4 A0A164ZVC2 A0A1I8MHA6 Q1HQJ6 A0A0P4VLD5 R4G4S2 A0A0P6BNG2 A0A0P6IKD8 A0A0A9Y776 A0A069DST6 A0A0K8TRE0 A0A2M4AQA9 A0A1A9ZBX2 A0A1S3IFV2 A0A2M4BTZ3 A0A023EQR6 A0A2M4BT24 A0A2M4CN88 A0A034VL69 A0A0V0GBV4 A0A023F854 A0A2M3ZII2 A0A2M3ZA66 W8BQT8 A0A0K8UII4 A0A1S3IFZ3 W5JAZ1 A0A224XNG1 A0A0A1X1D1 F4X079 E2AMC1 A0A1S3DQ43 A0A2J7QSZ6 A0A195F6M9 A0A1S3DPI1 A0A1Q3G4K2 E0VUI1 T1EA16 A0A195AZ03 A0A1W4UCG5 A0A1B6GBQ1 A0A1Q3G4N5 B3M7P5 B4IFZ2 A0A2S2NSU0 Q29DI9 A0A2H8TKP5 Q9VVW3
A0A1L8DZU1 A0A1L8DZU2 A0A1L8E039 A0A1B0CIK1 U5EUP8 A0A1W4X7R8 D7EIS6 A0A0N0BFF2 A0A1Y1M0T6 A0A067QH17 A0A088A4I8 A0A1J1HN73 J3JXU8 A0A2A3EEW6 A0A151JMQ4 A0A154PLW9 A0A151XDE3 A0A158NH46 A0A2J7QSX6 N6U2G2 K7INS3 A0A232FEJ9 A0A195D1C1 E9IMV8 A0A182V663 A0A182TWA3 A0A182KQE2 A0A182WWY6 Q7QIX5 A0A182I2I5 A0A182JND0 A0A182P0N0 A0A1A9XK37 R4WDH4 A0A182RP57 A0A2C9GW75 A0A182NK62 A0A182YEK7 A0A182SW02 A0A1I8PE01 A0A182WLI0 E9HAV9 A0A026X2I9 A0A182MNN6 A0A0T6B1A2 A0A1B6CGB4 A0A164ZVC2 A0A1I8MHA6 Q1HQJ6 A0A0P4VLD5 R4G4S2 A0A0P6BNG2 A0A0P6IKD8 A0A0A9Y776 A0A069DST6 A0A0K8TRE0 A0A2M4AQA9 A0A1A9ZBX2 A0A1S3IFV2 A0A2M4BTZ3 A0A023EQR6 A0A2M4BT24 A0A2M4CN88 A0A034VL69 A0A0V0GBV4 A0A023F854 A0A2M3ZII2 A0A2M3ZA66 W8BQT8 A0A0K8UII4 A0A1S3IFZ3 W5JAZ1 A0A224XNG1 A0A0A1X1D1 F4X079 E2AMC1 A0A1S3DQ43 A0A2J7QSZ6 A0A195F6M9 A0A1S3DPI1 A0A1Q3G4K2 E0VUI1 T1EA16 A0A195AZ03 A0A1W4UCG5 A0A1B6GBQ1 A0A1Q3G4N5 B3M7P5 B4IFZ2 A0A2S2NSU0 Q29DI9 A0A2H8TKP5 Q9VVW3
Pubmed
22118469
26354079
23674305
18362917
19820115
28004739
+ More
24845553 22516182 21347285 23537049 20075255 28648823 21282665 20966253 12364791 14747013 17210077 23691247 25244985 21292972 24508170 30249741 25315136 17204158 17510324 27129103 25401762 26823975 26334808 26369729 24945155 25348373 25474469 24495485 20920257 23761445 25830018 21719571 20798317 20566863 17994087 15632085 10731132 12537572 12537569 30442778
24845553 22516182 21347285 23537049 20075255 28648823 21282665 20966253 12364791 14747013 17210077 23691247 25244985 21292972 24508170 30249741 25315136 17204158 17510324 27129103 25401762 26823975 26334808 26369729 24945155 25348373 25474469 24495485 20920257 23761445 25830018 21719571 20798317 20566863 17994087 15632085 10731132 12537572 12537569 30442778
EMBL
NWSH01003869
PCG65741.1
AGBW02014495
OWR41578.1
KQ460973
KPJ10109.1
+ More
KQ459144 KPJ03754.1 GDQN01006850 JAT84204.1 KC469894 AGC92715.1 GFDF01002066 JAV12018.1 GFDF01002064 JAV12020.1 GFDF01002065 JAV12019.1 AJWK01013486 GANO01001372 JAB58499.1 KQ972823 EFA12262.2 KQ435800 KOX73268.1 GEZM01044388 JAV78260.1 KK853419 KDR07724.1 CVRI01000013 CRK89501.1 BT128070 AEE63031.1 KZ288266 PBC30277.1 KQ978899 KYN27614.1 KQ434948 KZC12444.1 KQ982294 KYQ58391.1 ADTU01015570 NEVH01011208 PNF31692.1 APGK01052195 KB741211 KB632263 ENN72747.1 ERL90880.1 NNAY01000323 OXU29214.1 KQ976973 KYN06710.1 GL764255 EFZ18095.1 AAAB01008807 EAA04211.4 APCN01000164 AK417708 BAN20923.1 AXCN02000428 GL732613 EFX71151.1 KK107020 QOIP01000006 EZA62502.1 RLU21946.1 AXCM01001237 LJIG01016243 KRT81187.1 GEDC01024869 GEDC01022250 GEDC01009862 GEDC01002033 JAS12429.1 JAS15048.1 JAS27436.1 JAS35265.1 LRGB01000687 KZS16819.1 DQ440448 CH477615 ABF18481.1 EAT38265.1 GDKW01001383 JAI55212.1 ACPB03015952 ACPB03015953 GAHY01001110 JAA76400.1 GDIP01012198 JAM91517.1 GDIQ01004087 JAN90650.1 GBHO01015595 GDHC01021912 GDHC01003788 JAG28009.1 JAP96716.1 JAQ14841.1 GBGD01002157 JAC86732.1 GDAI01001113 JAI16490.1 GGFK01009664 MBW42985.1 GGFJ01007415 MBW56556.1 GAPW01002632 JAC10966.1 GGFJ01007002 MBW56143.1 GGFL01002619 MBW66797.1 GAKP01016080 JAC42872.1 GECL01001261 JAP04863.1 GBBI01001231 JAC17481.1 GGFM01007626 MBW28377.1 GGFM01004619 MBW25370.1 GAMC01005129 GAMC01005128 JAC01428.1 GDHF01025963 GDHF01010967 JAI26351.1 JAI41347.1 ADMH02001644 ETN61627.1 GFTR01005068 JAW11358.1 GBXI01009836 GBXI01009143 JAD04456.1 JAD05149.1 GL888493 EGI60144.1 GL440811 EFN65412.1 PNF31693.1 KQ981768 KYN35847.1 GFDL01000318 JAV34727.1 AAZO01005495 DS235786 EEB17037.1 GAMD01000812 JAB00779.1 KQ976698 KYM77473.1 GECZ01022959 GECZ01009915 JAS46810.1 JAS59854.1 GFDL01000316 JAV34729.1 CH902618 EDV40973.1 CH480834 EDW46579.1 GGMR01007641 MBY20260.1 CH379070 EAL30425.3 GFXV01002901 MBW14706.1 AE014296 AY071029 BT044086
KQ459144 KPJ03754.1 GDQN01006850 JAT84204.1 KC469894 AGC92715.1 GFDF01002066 JAV12018.1 GFDF01002064 JAV12020.1 GFDF01002065 JAV12019.1 AJWK01013486 GANO01001372 JAB58499.1 KQ972823 EFA12262.2 KQ435800 KOX73268.1 GEZM01044388 JAV78260.1 KK853419 KDR07724.1 CVRI01000013 CRK89501.1 BT128070 AEE63031.1 KZ288266 PBC30277.1 KQ978899 KYN27614.1 KQ434948 KZC12444.1 KQ982294 KYQ58391.1 ADTU01015570 NEVH01011208 PNF31692.1 APGK01052195 KB741211 KB632263 ENN72747.1 ERL90880.1 NNAY01000323 OXU29214.1 KQ976973 KYN06710.1 GL764255 EFZ18095.1 AAAB01008807 EAA04211.4 APCN01000164 AK417708 BAN20923.1 AXCN02000428 GL732613 EFX71151.1 KK107020 QOIP01000006 EZA62502.1 RLU21946.1 AXCM01001237 LJIG01016243 KRT81187.1 GEDC01024869 GEDC01022250 GEDC01009862 GEDC01002033 JAS12429.1 JAS15048.1 JAS27436.1 JAS35265.1 LRGB01000687 KZS16819.1 DQ440448 CH477615 ABF18481.1 EAT38265.1 GDKW01001383 JAI55212.1 ACPB03015952 ACPB03015953 GAHY01001110 JAA76400.1 GDIP01012198 JAM91517.1 GDIQ01004087 JAN90650.1 GBHO01015595 GDHC01021912 GDHC01003788 JAG28009.1 JAP96716.1 JAQ14841.1 GBGD01002157 JAC86732.1 GDAI01001113 JAI16490.1 GGFK01009664 MBW42985.1 GGFJ01007415 MBW56556.1 GAPW01002632 JAC10966.1 GGFJ01007002 MBW56143.1 GGFL01002619 MBW66797.1 GAKP01016080 JAC42872.1 GECL01001261 JAP04863.1 GBBI01001231 JAC17481.1 GGFM01007626 MBW28377.1 GGFM01004619 MBW25370.1 GAMC01005129 GAMC01005128 JAC01428.1 GDHF01025963 GDHF01010967 JAI26351.1 JAI41347.1 ADMH02001644 ETN61627.1 GFTR01005068 JAW11358.1 GBXI01009836 GBXI01009143 JAD04456.1 JAD05149.1 GL888493 EGI60144.1 GL440811 EFN65412.1 PNF31693.1 KQ981768 KYN35847.1 GFDL01000318 JAV34727.1 AAZO01005495 DS235786 EEB17037.1 GAMD01000812 JAB00779.1 KQ976698 KYM77473.1 GECZ01022959 GECZ01009915 JAS46810.1 JAS59854.1 GFDL01000316 JAV34729.1 CH902618 EDV40973.1 CH480834 EDW46579.1 GGMR01007641 MBY20260.1 CH379070 EAL30425.3 GFXV01002901 MBW14706.1 AE014296 AY071029 BT044086
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000092461
UP000192223
+ More
UP000007266 UP000053105 UP000027135 UP000005203 UP000183832 UP000242457 UP000078492 UP000076502 UP000075809 UP000005205 UP000235965 UP000019118 UP000030742 UP000002358 UP000215335 UP000078542 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075885 UP000092443 UP000075900 UP000075886 UP000075884 UP000076408 UP000075901 UP000095300 UP000075920 UP000000305 UP000053097 UP000279307 UP000075883 UP000076858 UP000095301 UP000008820 UP000015103 UP000092445 UP000085678 UP000000673 UP000007755 UP000000311 UP000079169 UP000078541 UP000009046 UP000078540 UP000192221 UP000007801 UP000001292 UP000001819 UP000000803
UP000007266 UP000053105 UP000027135 UP000005203 UP000183832 UP000242457 UP000078492 UP000076502 UP000075809 UP000005205 UP000235965 UP000019118 UP000030742 UP000002358 UP000215335 UP000078542 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075885 UP000092443 UP000075900 UP000075886 UP000075884 UP000076408 UP000075901 UP000095300 UP000075920 UP000000305 UP000053097 UP000279307 UP000075883 UP000076858 UP000095301 UP000008820 UP000015103 UP000092445 UP000085678 UP000000673 UP000007755 UP000000311 UP000079169 UP000078541 UP000009046 UP000078540 UP000192221 UP000007801 UP000001292 UP000001819 UP000000803
PRIDE
ProteinModelPortal
A0A2A4J177
A0A212EJE5
A0A0N1IE59
A0A194QEA8
A0A1E1WB48
L7X3Y2
+ More
A0A1L8DZU1 A0A1L8DZU2 A0A1L8E039 A0A1B0CIK1 U5EUP8 A0A1W4X7R8 D7EIS6 A0A0N0BFF2 A0A1Y1M0T6 A0A067QH17 A0A088A4I8 A0A1J1HN73 J3JXU8 A0A2A3EEW6 A0A151JMQ4 A0A154PLW9 A0A151XDE3 A0A158NH46 A0A2J7QSX6 N6U2G2 K7INS3 A0A232FEJ9 A0A195D1C1 E9IMV8 A0A182V663 A0A182TWA3 A0A182KQE2 A0A182WWY6 Q7QIX5 A0A182I2I5 A0A182JND0 A0A182P0N0 A0A1A9XK37 R4WDH4 A0A182RP57 A0A2C9GW75 A0A182NK62 A0A182YEK7 A0A182SW02 A0A1I8PE01 A0A182WLI0 E9HAV9 A0A026X2I9 A0A182MNN6 A0A0T6B1A2 A0A1B6CGB4 A0A164ZVC2 A0A1I8MHA6 Q1HQJ6 A0A0P4VLD5 R4G4S2 A0A0P6BNG2 A0A0P6IKD8 A0A0A9Y776 A0A069DST6 A0A0K8TRE0 A0A2M4AQA9 A0A1A9ZBX2 A0A1S3IFV2 A0A2M4BTZ3 A0A023EQR6 A0A2M4BT24 A0A2M4CN88 A0A034VL69 A0A0V0GBV4 A0A023F854 A0A2M3ZII2 A0A2M3ZA66 W8BQT8 A0A0K8UII4 A0A1S3IFZ3 W5JAZ1 A0A224XNG1 A0A0A1X1D1 F4X079 E2AMC1 A0A1S3DQ43 A0A2J7QSZ6 A0A195F6M9 A0A1S3DPI1 A0A1Q3G4K2 E0VUI1 T1EA16 A0A195AZ03 A0A1W4UCG5 A0A1B6GBQ1 A0A1Q3G4N5 B3M7P5 B4IFZ2 A0A2S2NSU0 Q29DI9 A0A2H8TKP5 Q9VVW3
A0A1L8DZU1 A0A1L8DZU2 A0A1L8E039 A0A1B0CIK1 U5EUP8 A0A1W4X7R8 D7EIS6 A0A0N0BFF2 A0A1Y1M0T6 A0A067QH17 A0A088A4I8 A0A1J1HN73 J3JXU8 A0A2A3EEW6 A0A151JMQ4 A0A154PLW9 A0A151XDE3 A0A158NH46 A0A2J7QSX6 N6U2G2 K7INS3 A0A232FEJ9 A0A195D1C1 E9IMV8 A0A182V663 A0A182TWA3 A0A182KQE2 A0A182WWY6 Q7QIX5 A0A182I2I5 A0A182JND0 A0A182P0N0 A0A1A9XK37 R4WDH4 A0A182RP57 A0A2C9GW75 A0A182NK62 A0A182YEK7 A0A182SW02 A0A1I8PE01 A0A182WLI0 E9HAV9 A0A026X2I9 A0A182MNN6 A0A0T6B1A2 A0A1B6CGB4 A0A164ZVC2 A0A1I8MHA6 Q1HQJ6 A0A0P4VLD5 R4G4S2 A0A0P6BNG2 A0A0P6IKD8 A0A0A9Y776 A0A069DST6 A0A0K8TRE0 A0A2M4AQA9 A0A1A9ZBX2 A0A1S3IFV2 A0A2M4BTZ3 A0A023EQR6 A0A2M4BT24 A0A2M4CN88 A0A034VL69 A0A0V0GBV4 A0A023F854 A0A2M3ZII2 A0A2M3ZA66 W8BQT8 A0A0K8UII4 A0A1S3IFZ3 W5JAZ1 A0A224XNG1 A0A0A1X1D1 F4X079 E2AMC1 A0A1S3DQ43 A0A2J7QSZ6 A0A195F6M9 A0A1S3DPI1 A0A1Q3G4K2 E0VUI1 T1EA16 A0A195AZ03 A0A1W4UCG5 A0A1B6GBQ1 A0A1Q3G4N5 B3M7P5 B4IFZ2 A0A2S2NSU0 Q29DI9 A0A2H8TKP5 Q9VVW3
Ontologies
GO
PANTHER
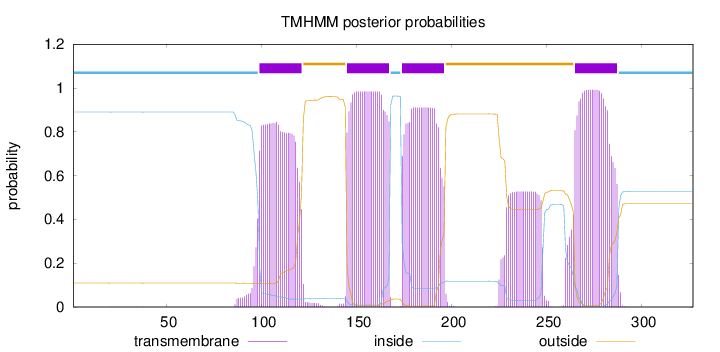
Topology
Subcellular location
Mitochondrion membrane
Length:
327
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
94.1585500000001
Exp number, first 60 AAs:
0.01209
Total prob of N-in:
0.89055
inside
1 - 98
TMhelix
99 - 121
outside
122 - 144
TMhelix
145 - 167
inside
168 - 173
TMhelix
174 - 196
outside
197 - 264
TMhelix
265 - 287
inside
288 - 327
Population Genetic Test Statistics
Pi
220.569083
Theta
183.701093
Tajima's D
0.713135
CLR
0.127034
CSRT
0.576821158942053
Interpretation
Uncertain
