Pre Gene Modal
BGIBMGA011296
Annotation
PREDICTED:_UPF0562_protein_C7orf55_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.335
Sequence
CDS
ATGGCATCCATCACTTCGAAGCCCGTGCTTGTTACACTGAGGCAGCTATTATCCGAACTTCGTAGGCAGAGTCCAACGAAGAAGTTAGCTGAAAATCAAATGGTGAGGTACATATTTGACCAGTACCGTAAATACCAGACCACAGATCAGCAATTGTGTAAGGCTATTGATGAAATGCACTTCCGAGCAAGAACTTATCATAATTATTTGCATCACTCGAGGAAATATAAAGAACTCAACACTGAGTTTAAAGGAAAGGGTGAGAGAAGTATTGAAGAGACAGCAAATATGGTCGGATTTAAGCTGCCTCATGACCCGAAGCCATAA
Protein
MASITSKPVLVTLRQLLSELRRQSPTKKLAENQMVRYIFDQYRKYQTTDQQLCKAIDEMHFRARTYHNYLHHSRKYKELNTEFKGKGERSIEETANMVGFKLPHDPKP
Summary
Uniprot
A0A2A4JLP8
A0A2H1WIH6
A0A0L7KWC6
A0A194QE53
S4P840
A0A0N1ICD7
+ More
A0A1Y1L668 A0A1I8MRJ0 U4UGB9 N6T473 A0A336MEE9 A0A1L8E8M2 A0A1L8E8V1 A0A1B0CCV0 A0A0K8SMV8 A0A1L8DB05 A0A1L8DBA3 B4K4Z0 A0A1A9WPC0 A0A146MES5 B4GP53 A0A1L8DB86 A0A1A9Z281 A0A1L8DBC6 B5DYD6 A0A0A1XCV9 W8C6R2 A0A034WT18 A0A2J7PCR6 B4NAI5 A0A0M4F5I2 A0A1B0G9N7 B3M066 B4PUM6 A0A1W4WWV8 A0A0N8P1C3 A0A1A9Y8T8 A0A3B0JLG1 B4HKE8 A0A1W4W2C2 Q0IGZ9 Q0KI97 A0A1L8D8M9 B3P1Q7 A0A067RHE2 B4QWN4 A0A2A3E3Y7 K7J8B5 A0A0L7QKH5 A0A026VWT6 A0A3S4QYC0 A0A158NIQ1 F4WDI1 A0A146M564 A0A162DAP4 A0A151J1E8 A0A0K8TPB1 A0A0P5KMW6 E9IZN0 A0A0P4XHI0 T1I1N7 A0A146L703
A0A1Y1L668 A0A1I8MRJ0 U4UGB9 N6T473 A0A336MEE9 A0A1L8E8M2 A0A1L8E8V1 A0A1B0CCV0 A0A0K8SMV8 A0A1L8DB05 A0A1L8DBA3 B4K4Z0 A0A1A9WPC0 A0A146MES5 B4GP53 A0A1L8DB86 A0A1A9Z281 A0A1L8DBC6 B5DYD6 A0A0A1XCV9 W8C6R2 A0A034WT18 A0A2J7PCR6 B4NAI5 A0A0M4F5I2 A0A1B0G9N7 B3M066 B4PUM6 A0A1W4WWV8 A0A0N8P1C3 A0A1A9Y8T8 A0A3B0JLG1 B4HKE8 A0A1W4W2C2 Q0IGZ9 Q0KI97 A0A1L8D8M9 B3P1Q7 A0A067RHE2 B4QWN4 A0A2A3E3Y7 K7J8B5 A0A0L7QKH5 A0A026VWT6 A0A3S4QYC0 A0A158NIQ1 F4WDI1 A0A146M564 A0A162DAP4 A0A151J1E8 A0A0K8TPB1 A0A0P5KMW6 E9IZN0 A0A0P4XHI0 T1I1N7 A0A146L703
Pubmed
EMBL
NWSH01001034
PCG73005.1
ODYU01008888
SOQ52868.1
JTDY01004943
KOB67572.1
+ More
KQ459144 KPJ03757.1 GAIX01006111 JAA86449.1 KQ460973 KPJ10112.1 GEZM01063260 GEZM01063259 JAV69162.1 KB632309 ERL92077.1 APGK01044524 KB741026 ENN74949.1 UFQS01000716 UFQS01000742 UFQT01000716 UFQT01000742 SSX06373.1 SSX06510.1 SSX26727.1 GFDG01003856 JAV14943.1 GFDG01003764 JAV15035.1 AJWK01007124 GBRD01011147 JAG54677.1 GFDF01010438 JAV03646.1 GFDF01010437 JAV03647.1 CH933806 EDW13961.1 GDHC01001403 JAQ17226.1 CH479186 EDW38936.1 GFDF01010358 JAV03726.1 GFDF01010321 JAV03763.1 CM000070 EDY67588.2 GBXI01005510 JAD08782.1 GAMC01008386 JAB98169.1 GAKP01002026 GAKP01002024 JAC56928.1 NEVH01027057 PNF14131.1 CH964232 EDW80799.1 CP012526 ALC47137.1 CCAG010009097 CH902617 EDV42023.1 CM000160 EDW96643.1 KPU79524.1 OUUW01000005 SPP81172.1 CH480815 EDW42898.1 BT028771 ABI34152.2 AE014297 KX531976 ABI31150.1 ANY27786.1 GFDF01011367 JAV02717.1 CH954181 EDV49795.1 KK852468 KDR23276.1 CM000364 EDX13638.1 KZ288422 PBC25966.1 AAZX01003472 KQ414940 KOC59127.1 KK107894 QOIP01000014 EZA47319.1 RLU15063.1 NCKU01002609 NCKU01002604 RWS09214.1 RWS09229.1 ADTU01017046 GL888087 EGI67761.1 GDHC01014351 GDHC01004214 JAQ04278.1 JAQ14415.1 LRGB01002101 KZS09204.1 KQ980522 KYN15684.1 GDAI01001610 JAI15993.1 GDIQ01207349 JAK44376.1 GL767232 EFZ13993.1 GDIP01242584 JAI80817.1 ACPB03005947 GDHC01015672 JAQ02957.1
KQ459144 KPJ03757.1 GAIX01006111 JAA86449.1 KQ460973 KPJ10112.1 GEZM01063260 GEZM01063259 JAV69162.1 KB632309 ERL92077.1 APGK01044524 KB741026 ENN74949.1 UFQS01000716 UFQS01000742 UFQT01000716 UFQT01000742 SSX06373.1 SSX06510.1 SSX26727.1 GFDG01003856 JAV14943.1 GFDG01003764 JAV15035.1 AJWK01007124 GBRD01011147 JAG54677.1 GFDF01010438 JAV03646.1 GFDF01010437 JAV03647.1 CH933806 EDW13961.1 GDHC01001403 JAQ17226.1 CH479186 EDW38936.1 GFDF01010358 JAV03726.1 GFDF01010321 JAV03763.1 CM000070 EDY67588.2 GBXI01005510 JAD08782.1 GAMC01008386 JAB98169.1 GAKP01002026 GAKP01002024 JAC56928.1 NEVH01027057 PNF14131.1 CH964232 EDW80799.1 CP012526 ALC47137.1 CCAG010009097 CH902617 EDV42023.1 CM000160 EDW96643.1 KPU79524.1 OUUW01000005 SPP81172.1 CH480815 EDW42898.1 BT028771 ABI34152.2 AE014297 KX531976 ABI31150.1 ANY27786.1 GFDF01011367 JAV02717.1 CH954181 EDV49795.1 KK852468 KDR23276.1 CM000364 EDX13638.1 KZ288422 PBC25966.1 AAZX01003472 KQ414940 KOC59127.1 KK107894 QOIP01000014 EZA47319.1 RLU15063.1 NCKU01002609 NCKU01002604 RWS09214.1 RWS09229.1 ADTU01017046 GL888087 EGI67761.1 GDHC01014351 GDHC01004214 JAQ04278.1 JAQ14415.1 LRGB01002101 KZS09204.1 KQ980522 KYN15684.1 GDAI01001610 JAI15993.1 GDIQ01207349 JAK44376.1 GL767232 EFZ13993.1 GDIP01242584 JAI80817.1 ACPB03005947 GDHC01015672 JAQ02957.1
Proteomes
UP000218220
UP000037510
UP000053268
UP000053240
UP000095301
UP000030742
+ More
UP000019118 UP000092461 UP000009192 UP000091820 UP000008744 UP000092445 UP000001819 UP000235965 UP000007798 UP000092553 UP000092444 UP000007801 UP000002282 UP000192223 UP000092443 UP000268350 UP000001292 UP000192221 UP000000803 UP000008711 UP000027135 UP000000304 UP000242457 UP000002358 UP000053825 UP000053097 UP000279307 UP000285301 UP000005205 UP000007755 UP000076858 UP000078492 UP000015103
UP000019118 UP000092461 UP000009192 UP000091820 UP000008744 UP000092445 UP000001819 UP000235965 UP000007798 UP000092553 UP000092444 UP000007801 UP000002282 UP000192223 UP000092443 UP000268350 UP000001292 UP000192221 UP000000803 UP000008711 UP000027135 UP000000304 UP000242457 UP000002358 UP000053825 UP000053097 UP000279307 UP000285301 UP000005205 UP000007755 UP000076858 UP000078492 UP000015103
ProteinModelPortal
A0A2A4JLP8
A0A2H1WIH6
A0A0L7KWC6
A0A194QE53
S4P840
A0A0N1ICD7
+ More
A0A1Y1L668 A0A1I8MRJ0 U4UGB9 N6T473 A0A336MEE9 A0A1L8E8M2 A0A1L8E8V1 A0A1B0CCV0 A0A0K8SMV8 A0A1L8DB05 A0A1L8DBA3 B4K4Z0 A0A1A9WPC0 A0A146MES5 B4GP53 A0A1L8DB86 A0A1A9Z281 A0A1L8DBC6 B5DYD6 A0A0A1XCV9 W8C6R2 A0A034WT18 A0A2J7PCR6 B4NAI5 A0A0M4F5I2 A0A1B0G9N7 B3M066 B4PUM6 A0A1W4WWV8 A0A0N8P1C3 A0A1A9Y8T8 A0A3B0JLG1 B4HKE8 A0A1W4W2C2 Q0IGZ9 Q0KI97 A0A1L8D8M9 B3P1Q7 A0A067RHE2 B4QWN4 A0A2A3E3Y7 K7J8B5 A0A0L7QKH5 A0A026VWT6 A0A3S4QYC0 A0A158NIQ1 F4WDI1 A0A146M564 A0A162DAP4 A0A151J1E8 A0A0K8TPB1 A0A0P5KMW6 E9IZN0 A0A0P4XHI0 T1I1N7 A0A146L703
A0A1Y1L668 A0A1I8MRJ0 U4UGB9 N6T473 A0A336MEE9 A0A1L8E8M2 A0A1L8E8V1 A0A1B0CCV0 A0A0K8SMV8 A0A1L8DB05 A0A1L8DBA3 B4K4Z0 A0A1A9WPC0 A0A146MES5 B4GP53 A0A1L8DB86 A0A1A9Z281 A0A1L8DBC6 B5DYD6 A0A0A1XCV9 W8C6R2 A0A034WT18 A0A2J7PCR6 B4NAI5 A0A0M4F5I2 A0A1B0G9N7 B3M066 B4PUM6 A0A1W4WWV8 A0A0N8P1C3 A0A1A9Y8T8 A0A3B0JLG1 B4HKE8 A0A1W4W2C2 Q0IGZ9 Q0KI97 A0A1L8D8M9 B3P1Q7 A0A067RHE2 B4QWN4 A0A2A3E3Y7 K7J8B5 A0A0L7QKH5 A0A026VWT6 A0A3S4QYC0 A0A158NIQ1 F4WDI1 A0A146M564 A0A162DAP4 A0A151J1E8 A0A0K8TPB1 A0A0P5KMW6 E9IZN0 A0A0P4XHI0 T1I1N7 A0A146L703
Ontologies
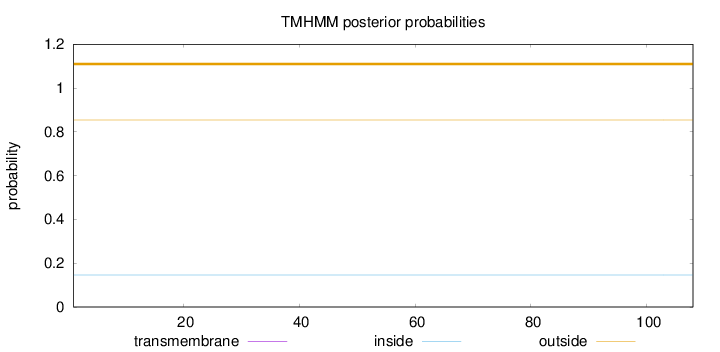
Topology
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14625
outside
1 - 108
Population Genetic Test Statistics
Pi
185.466098
Theta
189.357355
Tajima's D
-0.191627
CLR
1.885529
CSRT
0.322833858307085
Interpretation
Uncertain
