Gene
KWMTBOMO13923 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011308
Annotation
acyl-coenzyme_A_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.761 Mitochondrial Reliability : 1.255
Sequence
CDS
ATGGGCATCGAAACTCCGGTAGAATACAGCGGTTCCGGCTGCAATTTCCTGACAATGATGTTGGTGGTGGAAGAACTGTCTCGAGTGGACCCAGCCGTGGCCGCCTACGTCGACATTCACAACACCTTAGTCAACTCTCTGTTCATGAAGCTAGGTACAGAAGAACAAAAAAAGAAATATTTAACGAAACTCTGCACGGAATATGCTGGCAGTTTCTGTCTCACAGAGCCCAGCTCAGGATCAGATGCTTTTGCCTTGAAAACTGTTGCCAAAAAAGAAGGAGAACATTACATCATTAGCGGCTCTAAAATGTGGATCTCGAACTCTGATGTCGCTGGAGTCTTTCTAGTGATGACCAATGCTGATCCTTCCAAGGGTTACAAGGGCATCACCTGCTTTATCGTTGAACGTGAAACCCCGGGCCTCAGTGTTGCCAAGCCTGAAAACAAATTAGGCATCAGAGCTTCGGGCACATGCATGGTACATTTCGACAATGTGAAGGTGCACGAATCGAGTATCCTTGGAGAATATGGCAAAGGGTACAAGTACGCGGCTGGTTTCTTGAACGAAGGTCGGATTGGTATCGCGGCTCAGATGATCGGTCTGTGTCAGGGCTGTATGGACGCAACGATTCCTTACACACTAGAAAGGAAACAATTCGGAAAGAAGATTTACGAGTTTCAGGGCATCAGCTACCAAATCGCACACCTGCAGACACAGTTGGAAGCCGCCCGTCTTCTAACATACAACGCCGCTAGACTGAAAGAACACGGCCACGAGTTTGTTAAAGAAGCCGCGATGGCTAAATATTTTGCTTCAGAAATCGCACAAACGTTAACTTCAAAATGCATAGACTTTATGGGAGGCGTTGGTTTCACGAGAGATTTTCCGCAAGAAAAATTCTATAGGGACGCTAAAATTGGTACAATTTACGAGGGTACCAGCAACATGCAGTTACAAACGATTGCCAAATTATTAGAAAGGGAATACACACAATAA
Protein
MGIETPVEYSGSGCNFLTMMLVVEELSRVDPAVAAYVDIHNTLVNSLFMKLGTEEQKKKYLTKLCTEYAGSFCLTEPSSGSDAFALKTVAKKEGEHYIISGSKMWISNSDVAGVFLVMTNADPSKGYKGITCFIVERETPGLSVAKPENKLGIRASGTCMVHFDNVKVHESSILGEYGKGYKYAAGFLNEGRIGIAAQMIGLCQGCMDATIPYTLERKQFGKKIYEFQGISYQIAHLQTQLEAARLLTYNAARLKEHGHEFVKEAAMAKYFASEIAQTLTSKCIDFMGGVGFTRDFPQEKFYRDAKIGTIYEGTSNMQLQTIAKLLEREYTQ
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
Q19P04
H9JP54
A0A3S2TKV3
A0A068FKB7
A0A2H1WIL0
A0A2A4JN39
+ More
A0A0N1PFX2 I4DJW7 A0A194QDY8 S4PDU4 A0A034VXN0 A0A232FGL5 A0A182G6W1 K7J9F0 A0A0K8VCB6 A0A1B6H1P6 Q171S4 A0A0A1WUM1 T1D2H1 A0A1Q3EZA1 A0A1B6JQ28 A0A1Q3EZA4 A0A1L8DZL3 T1DGK4 B4N4V3 U5EQP8 A0A3B0JLC2 Q2M0E5 B4GRR0 W8C7I0 A0A1A9Z3D4 B4KZ04 A0A1I8QF43 B4LD52 A0A224XRK2 A0A1A9WFH3 A0A182IQS2 B0WQ26 A0A0P4VIT3 A0A069DT04 A0A1A9VYL2 A0A1A9XWJ0 A0A1B0AYA1 B4PH24 A0A0M4EDV0 A0A0V0G445 A0A084WG81 Q9VVU1 Q95U46 A0A1B6C654 D3TRT6 B4IYD8 W5JFB0 A0A182TMV4 A0A182KTZ0 A0A182XG95 A0A182HRU4 Q7Q1H8 B3M858 A0A1W4UP90 B3NDU9 A0A151X142 A0A1L8EFA1 A0A182UUR6 A0A151J1L4 A0A151I0M2 A0A182F2X3 A0A1B6CCZ7 A0A2M3Z2P1 A0A2M3ZGJ3 A0A1W4X7P8 A0A2M4BNT2 A0A182Q7G7 N6U8L7 A0A182P4C1 T1PBC4 A0A182JQ47 A0A2M4BP67 A0A088AFJ0 F4WDI2 A0A182YDE2 J3JUH9 A0A0J9RY01 A0A2M4A2S4 A0A2M4A328 A0A182MSQ2 A0A158NIP9 A0A154P2B0 A0A182NBU2 A0A2A3E2N0 A0A182SGH3 A0A336KJB0 A0A182REX5 A0A182WGL9 E2BRN5 A0A0A9Y8G7 A0A310S473 A0A0T6AYJ6 A0A195CBT9 A0A026VUH3
A0A0N1PFX2 I4DJW7 A0A194QDY8 S4PDU4 A0A034VXN0 A0A232FGL5 A0A182G6W1 K7J9F0 A0A0K8VCB6 A0A1B6H1P6 Q171S4 A0A0A1WUM1 T1D2H1 A0A1Q3EZA1 A0A1B6JQ28 A0A1Q3EZA4 A0A1L8DZL3 T1DGK4 B4N4V3 U5EQP8 A0A3B0JLC2 Q2M0E5 B4GRR0 W8C7I0 A0A1A9Z3D4 B4KZ04 A0A1I8QF43 B4LD52 A0A224XRK2 A0A1A9WFH3 A0A182IQS2 B0WQ26 A0A0P4VIT3 A0A069DT04 A0A1A9VYL2 A0A1A9XWJ0 A0A1B0AYA1 B4PH24 A0A0M4EDV0 A0A0V0G445 A0A084WG81 Q9VVU1 Q95U46 A0A1B6C654 D3TRT6 B4IYD8 W5JFB0 A0A182TMV4 A0A182KTZ0 A0A182XG95 A0A182HRU4 Q7Q1H8 B3M858 A0A1W4UP90 B3NDU9 A0A151X142 A0A1L8EFA1 A0A182UUR6 A0A151J1L4 A0A151I0M2 A0A182F2X3 A0A1B6CCZ7 A0A2M3Z2P1 A0A2M3ZGJ3 A0A1W4X7P8 A0A2M4BNT2 A0A182Q7G7 N6U8L7 A0A182P4C1 T1PBC4 A0A182JQ47 A0A2M4BP67 A0A088AFJ0 F4WDI2 A0A182YDE2 J3JUH9 A0A0J9RY01 A0A2M4A2S4 A0A2M4A328 A0A182MSQ2 A0A158NIP9 A0A154P2B0 A0A182NBU2 A0A2A3E2N0 A0A182SGH3 A0A336KJB0 A0A182REX5 A0A182WGL9 E2BRN5 A0A0A9Y8G7 A0A310S473 A0A0T6AYJ6 A0A195CBT9 A0A026VUH3
Pubmed
19121390
26385554
26354079
22651552
23622113
25348373
+ More
28648823 26483478 20075255 17510324 25830018 24330624 17994087 15632085 24495485 27129103 26334808 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20920257 23761445 20966253 12364791 23537049 25315136 21719571 25244985 22516182 22936249 21347285 20798317 25401762 26823975 24508170
28648823 26483478 20075255 17510324 25830018 24330624 17994087 15632085 24495485 27129103 26334808 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20920257 23761445 20966253 12364791 23537049 25315136 21719571 25244985 22516182 22936249 21347285 20798317 25401762 26823975 24508170
EMBL
DQ515927
ABF71566.1
BABH01009531
BABH01009532
BABH01009533
RSAL01000075
+ More
RVE48878.1 KJ622079 AID66669.1 ODYU01008888 SOQ52867.1 NWSH01001034 PCG73004.1 KQ460973 KPJ10113.1 AK401585 BAM18207.1 KQ459144 KPJ03758.1 GAIX01004807 JAA87753.1 GAKP01012327 JAC46625.1 NNAY01000224 OXU29884.1 JXUM01045772 KQ561450 KXJ78547.1 GDHF01015720 JAI36594.1 GECZ01008382 GECZ01001229 JAS61387.1 JAS68540.1 CH477447 EAT40743.1 GBXI01013553 GBXI01011543 JAD00739.1 JAD02749.1 GALA01001618 JAA93234.1 GFDL01014408 JAV20637.1 GECU01006387 JAT01320.1 GFDL01014414 JAV20631.1 GFDF01002369 JAV11715.1 GALA01001621 JAA93231.1 CH964101 EDW79392.1 GANO01003190 JAB56681.1 OUUW01000002 SPP76200.1 CH379069 EAL30989.2 CH479188 EDW40445.1 GAMC01003609 JAC02947.1 CH933809 EDW18896.1 CH940647 EDW69933.1 GFTR01005653 JAW10773.1 DS232032 EDS32670.1 GDKW01002663 JAI53932.1 GBGD01001809 JAC87080.1 JXJN01005581 CM000159 EDW95394.1 EDW95402.1 CP012525 ALC43834.1 GECL01003299 JAP02825.1 ATLV01023461 KE525343 KFB49225.1 AE014296 BT044429 AAF49216.1 ACH92494.1 AY058314 AAL13543.1 GEDC01028312 JAS08986.1 CCAG010023908 EZ424138 ADD20414.1 CH916366 EDV97611.1 ADMH02001647 ETN61570.1 APCN01001747 AAAB01008980 EAA14306.3 CH902618 EDV40997.1 CH954178 EDV52303.1 KQ982603 KYQ53976.1 GFDG01001397 JAV17402.1 KQ980522 KYN15683.1 KQ976601 KYM79433.1 GEDC01025962 JAS11336.1 GGFM01002036 MBW22787.1 GGFM01006915 MBW27666.1 GGFJ01005599 MBW54740.1 AXCN02000691 APGK01044519 KB741026 KB632309 ENN74947.1 ERL92079.1 KA645440 AFP60069.1 GGFJ01005600 MBW54741.1 GL888087 EGI67762.1 BT126893 AEE61855.1 CM002912 KMZ00463.1 GGFK01001782 MBW35103.1 GGFK01001799 MBW35120.1 AXCM01002017 ADTU01017045 ADTU01017046 KQ434803 KZC05997.1 KZ288422 PBC25967.1 UFQS01000514 UFQT01000514 SSX04527.1 SSX24890.1 GL449978 EFN81629.1 GBHO01040473 GBHO01015150 GBRD01010383 GDHC01013418 GDHC01006405 JAG03131.1 JAG28454.1 JAG55441.1 JAQ05211.1 JAQ12224.1 KQ780658 OAD51912.1 LJIG01022516 KRT80161.1 KQ978068 KYM97568.1 KK107894 EZA47320.1
RVE48878.1 KJ622079 AID66669.1 ODYU01008888 SOQ52867.1 NWSH01001034 PCG73004.1 KQ460973 KPJ10113.1 AK401585 BAM18207.1 KQ459144 KPJ03758.1 GAIX01004807 JAA87753.1 GAKP01012327 JAC46625.1 NNAY01000224 OXU29884.1 JXUM01045772 KQ561450 KXJ78547.1 GDHF01015720 JAI36594.1 GECZ01008382 GECZ01001229 JAS61387.1 JAS68540.1 CH477447 EAT40743.1 GBXI01013553 GBXI01011543 JAD00739.1 JAD02749.1 GALA01001618 JAA93234.1 GFDL01014408 JAV20637.1 GECU01006387 JAT01320.1 GFDL01014414 JAV20631.1 GFDF01002369 JAV11715.1 GALA01001621 JAA93231.1 CH964101 EDW79392.1 GANO01003190 JAB56681.1 OUUW01000002 SPP76200.1 CH379069 EAL30989.2 CH479188 EDW40445.1 GAMC01003609 JAC02947.1 CH933809 EDW18896.1 CH940647 EDW69933.1 GFTR01005653 JAW10773.1 DS232032 EDS32670.1 GDKW01002663 JAI53932.1 GBGD01001809 JAC87080.1 JXJN01005581 CM000159 EDW95394.1 EDW95402.1 CP012525 ALC43834.1 GECL01003299 JAP02825.1 ATLV01023461 KE525343 KFB49225.1 AE014296 BT044429 AAF49216.1 ACH92494.1 AY058314 AAL13543.1 GEDC01028312 JAS08986.1 CCAG010023908 EZ424138 ADD20414.1 CH916366 EDV97611.1 ADMH02001647 ETN61570.1 APCN01001747 AAAB01008980 EAA14306.3 CH902618 EDV40997.1 CH954178 EDV52303.1 KQ982603 KYQ53976.1 GFDG01001397 JAV17402.1 KQ980522 KYN15683.1 KQ976601 KYM79433.1 GEDC01025962 JAS11336.1 GGFM01002036 MBW22787.1 GGFM01006915 MBW27666.1 GGFJ01005599 MBW54740.1 AXCN02000691 APGK01044519 KB741026 KB632309 ENN74947.1 ERL92079.1 KA645440 AFP60069.1 GGFJ01005600 MBW54741.1 GL888087 EGI67762.1 BT126893 AEE61855.1 CM002912 KMZ00463.1 GGFK01001782 MBW35103.1 GGFK01001799 MBW35120.1 AXCM01002017 ADTU01017045 ADTU01017046 KQ434803 KZC05997.1 KZ288422 PBC25967.1 UFQS01000514 UFQT01000514 SSX04527.1 SSX24890.1 GL449978 EFN81629.1 GBHO01040473 GBHO01015150 GBRD01010383 GDHC01013418 GDHC01006405 JAG03131.1 JAG28454.1 JAG55441.1 JAQ05211.1 JAQ12224.1 KQ780658 OAD51912.1 LJIG01022516 KRT80161.1 KQ978068 KYM97568.1 KK107894 EZA47320.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000215335
+ More
UP000069940 UP000249989 UP000002358 UP000008820 UP000007798 UP000268350 UP000001819 UP000008744 UP000092445 UP000009192 UP000095300 UP000008792 UP000091820 UP000075880 UP000002320 UP000078200 UP000092443 UP000092460 UP000002282 UP000092553 UP000030765 UP000000803 UP000092444 UP000001070 UP000000673 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000007801 UP000192221 UP000008711 UP000075809 UP000075903 UP000078492 UP000078540 UP000069272 UP000192223 UP000075886 UP000019118 UP000030742 UP000075885 UP000095301 UP000075881 UP000005203 UP000007755 UP000076408 UP000075883 UP000005205 UP000076502 UP000075884 UP000242457 UP000075901 UP000075900 UP000075920 UP000008237 UP000078542 UP000053097
UP000069940 UP000249989 UP000002358 UP000008820 UP000007798 UP000268350 UP000001819 UP000008744 UP000092445 UP000009192 UP000095300 UP000008792 UP000091820 UP000075880 UP000002320 UP000078200 UP000092443 UP000092460 UP000002282 UP000092553 UP000030765 UP000000803 UP000092444 UP000001070 UP000000673 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000007801 UP000192221 UP000008711 UP000075809 UP000075903 UP000078492 UP000078540 UP000069272 UP000192223 UP000075886 UP000019118 UP000030742 UP000075885 UP000095301 UP000075881 UP000005203 UP000007755 UP000076408 UP000075883 UP000005205 UP000076502 UP000075884 UP000242457 UP000075901 UP000075900 UP000075920 UP000008237 UP000078542 UP000053097
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q19P04
H9JP54
A0A3S2TKV3
A0A068FKB7
A0A2H1WIL0
A0A2A4JN39
+ More
A0A0N1PFX2 I4DJW7 A0A194QDY8 S4PDU4 A0A034VXN0 A0A232FGL5 A0A182G6W1 K7J9F0 A0A0K8VCB6 A0A1B6H1P6 Q171S4 A0A0A1WUM1 T1D2H1 A0A1Q3EZA1 A0A1B6JQ28 A0A1Q3EZA4 A0A1L8DZL3 T1DGK4 B4N4V3 U5EQP8 A0A3B0JLC2 Q2M0E5 B4GRR0 W8C7I0 A0A1A9Z3D4 B4KZ04 A0A1I8QF43 B4LD52 A0A224XRK2 A0A1A9WFH3 A0A182IQS2 B0WQ26 A0A0P4VIT3 A0A069DT04 A0A1A9VYL2 A0A1A9XWJ0 A0A1B0AYA1 B4PH24 A0A0M4EDV0 A0A0V0G445 A0A084WG81 Q9VVU1 Q95U46 A0A1B6C654 D3TRT6 B4IYD8 W5JFB0 A0A182TMV4 A0A182KTZ0 A0A182XG95 A0A182HRU4 Q7Q1H8 B3M858 A0A1W4UP90 B3NDU9 A0A151X142 A0A1L8EFA1 A0A182UUR6 A0A151J1L4 A0A151I0M2 A0A182F2X3 A0A1B6CCZ7 A0A2M3Z2P1 A0A2M3ZGJ3 A0A1W4X7P8 A0A2M4BNT2 A0A182Q7G7 N6U8L7 A0A182P4C1 T1PBC4 A0A182JQ47 A0A2M4BP67 A0A088AFJ0 F4WDI2 A0A182YDE2 J3JUH9 A0A0J9RY01 A0A2M4A2S4 A0A2M4A328 A0A182MSQ2 A0A158NIP9 A0A154P2B0 A0A182NBU2 A0A2A3E2N0 A0A182SGH3 A0A336KJB0 A0A182REX5 A0A182WGL9 E2BRN5 A0A0A9Y8G7 A0A310S473 A0A0T6AYJ6 A0A195CBT9 A0A026VUH3
A0A0N1PFX2 I4DJW7 A0A194QDY8 S4PDU4 A0A034VXN0 A0A232FGL5 A0A182G6W1 K7J9F0 A0A0K8VCB6 A0A1B6H1P6 Q171S4 A0A0A1WUM1 T1D2H1 A0A1Q3EZA1 A0A1B6JQ28 A0A1Q3EZA4 A0A1L8DZL3 T1DGK4 B4N4V3 U5EQP8 A0A3B0JLC2 Q2M0E5 B4GRR0 W8C7I0 A0A1A9Z3D4 B4KZ04 A0A1I8QF43 B4LD52 A0A224XRK2 A0A1A9WFH3 A0A182IQS2 B0WQ26 A0A0P4VIT3 A0A069DT04 A0A1A9VYL2 A0A1A9XWJ0 A0A1B0AYA1 B4PH24 A0A0M4EDV0 A0A0V0G445 A0A084WG81 Q9VVU1 Q95U46 A0A1B6C654 D3TRT6 B4IYD8 W5JFB0 A0A182TMV4 A0A182KTZ0 A0A182XG95 A0A182HRU4 Q7Q1H8 B3M858 A0A1W4UP90 B3NDU9 A0A151X142 A0A1L8EFA1 A0A182UUR6 A0A151J1L4 A0A151I0M2 A0A182F2X3 A0A1B6CCZ7 A0A2M3Z2P1 A0A2M3ZGJ3 A0A1W4X7P8 A0A2M4BNT2 A0A182Q7G7 N6U8L7 A0A182P4C1 T1PBC4 A0A182JQ47 A0A2M4BP67 A0A088AFJ0 F4WDI2 A0A182YDE2 J3JUH9 A0A0J9RY01 A0A2M4A2S4 A0A2M4A328 A0A182MSQ2 A0A158NIP9 A0A154P2B0 A0A182NBU2 A0A2A3E2N0 A0A182SGH3 A0A336KJB0 A0A182REX5 A0A182WGL9 E2BRN5 A0A0A9Y8G7 A0A310S473 A0A0T6AYJ6 A0A195CBT9 A0A026VUH3
PDB
2JIF
E-value=4.62272e-120,
Score=1102
Ontologies
PATHWAY
GO
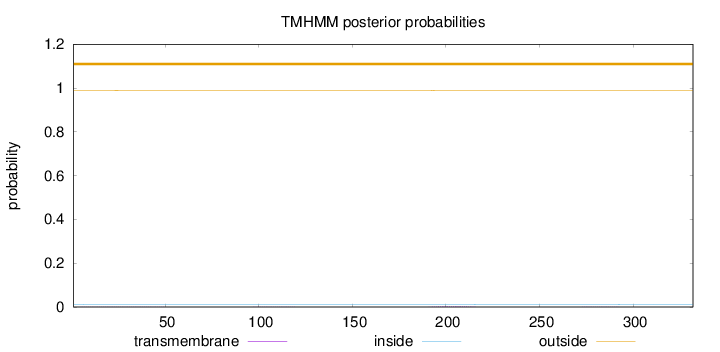
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0829699999999999
Exp number, first 60 AAs:
0.01708
Total prob of N-in:
0.01076
outside
1 - 332
Population Genetic Test Statistics
Pi
247.816198
Theta
188.276227
Tajima's D
2.059937
CLR
0.115181
CSRT
0.889405529723514
Interpretation
Uncertain
