Gene
KWMTBOMO13922 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011297
Annotation
activator_of_90_kDa_heat_shock_protein_ATPase_homolog_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.277
Sequence
CDS
ATGGCCAAGTGGGGCGAAGGTGACCCACGATGGATTGTGGAAGAGCGCCCGGACGCAACAAATGTGAACAACTGGCATTGGACGGAGAAAAACGCCGGCCCGTGGTCGAAAGATCGATTGAAAGAACTTTTCAGTGATTTGAAAATAGCTCAAAACGGCATTGTCTGTAGTATTACTGAAGTAGAAAAAGTCGACGGCGAAGCATCAGCGAACAATAGAAAAGGAAAATTAATATTTTTCTATGAATGGGACATAAAGTTGAAATGGGAAGGTGTGTTGGCTGGAGGTAGTGAAAAAATCAAGGGAGAGATTCACATCCCGAACTTATCCGAAGAAAATGACGTCAGTGAAGTGGACATGACAGTTACAATTAAAAGTAACGGAGATGAAGCCCAGAGAGTGAAAGCATTTATGCATCATGTAGGGCGGGAAGAGATCCGAAAACAACTCCAAGAGTATATTCGAAGTCTAAAAGAAGAGTTCTCAAAAGGCCTCATTCTGCCAAAGAAAGGCGAATCTTCCGTCAAACCTGACAATGTATCAACAATCACAAGTGGATTTAATAAAAAAATCAATATGAACCCAATAATCTCACCACAGACTAATAAAGTTGGCTGTAAACTGGATACAAAGACAATTGAACTATCAGAGAAATTTCAATGTCGAGGACAAGAATTTTATGATGCTATGACTAGAATAGAGATGGTGACTGCCTTCACTCAGGGTCATGTAAAGCTAGATCCCGAGAAAGGTGGAAAATTTGCCTTATTTGGTGGTAATGTTACTGGAGAATTTAAAGAACTCGTCCCAGGAAAGAGAATCGTTCAATATTGGAGATATAAGCAATGGCCTGAACAACACTACTCAGAAGTGACATTTAATATTGAAGAGAAGGATGACCACACTCTAGTCCGACTCAAACAAGACCTTGTGCCTGTCGCTGAGGTTGAGAAGACCCGTGACAACTGGCAACGATACTACTTTGACAGTATTAAGCGTGCTTTCGGATTTGGAGCGTTCCTGTGA
Protein
MAKWGEGDPRWIVEERPDATNVNNWHWTEKNAGPWSKDRLKELFSDLKIAQNGIVCSITEVEKVDGEASANNRKGKLIFFYEWDIKLKWEGVLAGGSEKIKGEIHIPNLSEENDVSEVDMTVTIKSNGDEAQRVKAFMHHVGREEIRKQLQEYIRSLKEEFSKGLILPKKGESSVKPDNVSTITSGFNKKINMNPIISPQTNKVGCKLDTKTIELSEKFQCRGQEFYDAMTRIEMVTAFTQGHVKLDPEKGGKFALFGGNVTGEFKELVPGKRIVQYWRYKQWPEQHYSEVTFNIEEKDDHTLVRLKQDLVPVAEVEKTRDNWQRYYFDSIKRAFGFGAFL
Summary
Uniprot
H9JP43
Q75RW2
A0A194QEB3
A0A2A4JNJ0
A0A2H1WIS9
A0A212F829
+ More
A0A0N1I5N0 S4P8R4 I4DP00 D6WCK9 U5EP77 A0A182G5K0 A0A023ERF3 A0A1Q3FWL3 Q178H4 A0A0L7LTJ4 A0A336LKJ5 W5JIF2 A0A1B0CUN5 A0A182WUU7 Q7PPS8 A0A182HFV6 A0A182V963 A0A182LPY7 A0A1L8DUC6 A0A084WAX6 A0A182K5S0 A0A1L8DUE3 A0A182QAQ2 A0A182RF43 A0A182MLW4 A0A2M4BSQ1 A0A2M4ANB2 A0A182JM66 A0A182FTR9 A0A069DS64 N6TU58 J3JU76 F4W8S2 A0A1Y9H2K6 A0A2M4ANE3 A0A158NKG6 A0A195FCI4 A0A2M3Z9X2 A0A224XRE9 A0A195B058 A0A0M8ZQ82 A0A182VT72 A0A088AJL2 A0A2A3E6P2 V9IL20 A0A0L7RE91 A0A1W4WAD1 A0A067R5R7 A0A0J7L5B0 A0A0P4VUN4 T1IC42 A0A026X249 R4WJL5 A0A232EXF5 E2B301 K7IUI9 A0A1B6EHD5 A0A1B6MUF2 A0A097PHM9 A0A1Y1LE46 E9IWZ4 Q9V9Q4 A0A1B6E8U8 A0A195EIL0 A0A0M5IWS2 A0A1A9ZR92 B4P6G1 B4N7K6 B3MTZ5 A0A1W4V286 D3TPD4 B4Q4F9 A0A1A9UWA3 B4IIG7 B3NKR7 B4JE28 A0A0A9XP10 A0A034WKD6 A0A0K8U2T3 A0A0A1WFN8 A0A3B0KVW5 Q29PF6 B4GKP1 A0A293LPH8 A0A3B0KLN3 A0A1A9YJL3 A0A182YF80 E0VNP2 A0A182PWG7 C4WRP4 A0A1B0ARP8 A0A182U942 A0A0C9SE15 A0A1E1XFF7
A0A0N1I5N0 S4P8R4 I4DP00 D6WCK9 U5EP77 A0A182G5K0 A0A023ERF3 A0A1Q3FWL3 Q178H4 A0A0L7LTJ4 A0A336LKJ5 W5JIF2 A0A1B0CUN5 A0A182WUU7 Q7PPS8 A0A182HFV6 A0A182V963 A0A182LPY7 A0A1L8DUC6 A0A084WAX6 A0A182K5S0 A0A1L8DUE3 A0A182QAQ2 A0A182RF43 A0A182MLW4 A0A2M4BSQ1 A0A2M4ANB2 A0A182JM66 A0A182FTR9 A0A069DS64 N6TU58 J3JU76 F4W8S2 A0A1Y9H2K6 A0A2M4ANE3 A0A158NKG6 A0A195FCI4 A0A2M3Z9X2 A0A224XRE9 A0A195B058 A0A0M8ZQ82 A0A182VT72 A0A088AJL2 A0A2A3E6P2 V9IL20 A0A0L7RE91 A0A1W4WAD1 A0A067R5R7 A0A0J7L5B0 A0A0P4VUN4 T1IC42 A0A026X249 R4WJL5 A0A232EXF5 E2B301 K7IUI9 A0A1B6EHD5 A0A1B6MUF2 A0A097PHM9 A0A1Y1LE46 E9IWZ4 Q9V9Q4 A0A1B6E8U8 A0A195EIL0 A0A0M5IWS2 A0A1A9ZR92 B4P6G1 B4N7K6 B3MTZ5 A0A1W4V286 D3TPD4 B4Q4F9 A0A1A9UWA3 B4IIG7 B3NKR7 B4JE28 A0A0A9XP10 A0A034WKD6 A0A0K8U2T3 A0A0A1WFN8 A0A3B0KVW5 Q29PF6 B4GKP1 A0A293LPH8 A0A3B0KLN3 A0A1A9YJL3 A0A182YF80 E0VNP2 A0A182PWG7 C4WRP4 A0A1B0ARP8 A0A182U942 A0A0C9SE15 A0A1E1XFF7
Pubmed
19121390
26354079
22118469
23622113
22651552
18362917
+ More
19820115 26483478 24945155 17510324 26227816 20920257 23761445 12364791 14747013 17210077 20966253 24438588 26334808 23537049 22516182 21719571 21347285 24845553 27129103 24508170 30249741 23691247 28648823 20798317 20075255 28004739 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20353571 22936249 25401762 26823975 25348373 25830018 15632085 25244985 20566863 26131772 28503490
19820115 26483478 24945155 17510324 26227816 20920257 23761445 12364791 14747013 17210077 20966253 24438588 26334808 23537049 22516182 21719571 21347285 24845553 27129103 24508170 30249741 23691247 28648823 20798317 20075255 28004739 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20353571 22936249 25401762 26823975 25348373 25830018 15632085 25244985 20566863 26131772 28503490
EMBL
BABH01009527
AB158647
BAD07028.1
KQ459144
KPJ03759.1
NWSH01001034
+ More
PCG73003.1 ODYU01008888 SOQ52866.1 AGBW02009804 OWR49890.1 KQ460973 KPJ10114.1 GAIX01004079 JAA88481.1 AK403221 BAM19640.1 KQ971311 EEZ99044.1 GANO01000277 JAB59594.1 JXUM01146283 KQ570143 KXJ68444.1 GAPW01002362 JAC11236.1 GFDL01003034 JAV32011.1 CH477363 EAT42609.1 EAT42610.1 EAT42611.1 JTDY01000154 KOB78561.1 UFQS01000039 UFQT01000039 SSW98090.1 SSX18476.1 ADMH02001074 ETN64167.1 AJWK01029436 AJWK01029437 AJWK01029438 AAAB01008933 EAA09900.4 APCN01005588 GFDF01004036 JAV10048.1 ATLV01022275 KE525331 KFB47370.1 GFDF01004035 JAV10049.1 AXCN02002042 AXCM01001028 GGFJ01006975 MBW56116.1 GGFK01008926 MBW42247.1 GBGD01002011 JAC86878.1 APGK01054399 APGK01054400 KB741249 ENN71926.1 BT126789 KB631760 AEE61751.1 ERL85899.1 GL887908 EGI69563.1 GGFK01008978 MBW42299.1 ADTU01018832 KQ981693 KYN37754.1 GGFM01004586 MBW25337.1 GFTR01005304 JAW11122.1 KQ976692 KYM77841.1 KQ436240 KOX67349.1 KZ288349 PBC27417.1 JR050887 AEY61377.1 KQ414612 KOC69144.1 KK852726 KDR17659.1 LBMM01000528 KMQ98132.1 GDKW01000330 JAI56265.1 ACPB03017890 KK107021 QOIP01000011 EZA62395.1 RLU17077.1 AK417755 BAN20970.1 NNAY01001765 OXU22977.1 GL445250 EFN89970.1 AAZX01004049 GECZ01032448 GECZ01011988 GECZ01009674 JAS37321.1 JAS57781.1 JAS60095.1 GEBQ01000403 JAT39574.1 KM221889 AIU47030.1 GEZM01058077 JAV71922.1 GL766616 EFZ14926.1 AE014134 AY121650 AAF57232.2 AAM51977.1 GEDC01002940 JAS34358.1 KQ978881 KYN27699.1 CP012523 ALC39831.1 CM000158 EDW89918.1 CH964214 EDW80345.1 CH902624 EDV33324.1 EZ423286 ADD19562.1 CM000361 CM002910 EDX05756.1 KMY91385.1 CH480843 EDW49717.1 CH954179 EDV54371.1 CH916368 EDW03548.1 GBHO01036467 GBHO01035468 GBHO01030840 GBHO01021132 GBHO01021131 GBHO01021129 GBHO01021126 GBHO01021125 GBHO01021124 GBRD01017077 GBRD01017074 GDHC01021752 GDHC01012825 JAG07137.1 JAG08136.1 JAG12764.1 JAG22472.1 JAG22473.1 JAG22475.1 JAG22478.1 JAG22479.1 JAG22480.1 JAG48750.1 JAP96876.1 JAQ05804.1 GAKP01002901 JAC56051.1 GDHF01031245 GDHF01017939 GDHF01017218 JAI21069.1 JAI34375.1 JAI35096.1 GBXI01016493 JAC97798.1 OUUW01000022 SPP89531.1 CH379058 EAL34337.2 CH479184 EDW37207.1 GFWV01005883 MAA30613.1 SPP89530.1 DS235341 EEB14998.1 AK339856 BAH70564.1 JXJN01002512 GBZX01001487 JAG91253.1 GFAC01001174 JAT98014.1
PCG73003.1 ODYU01008888 SOQ52866.1 AGBW02009804 OWR49890.1 KQ460973 KPJ10114.1 GAIX01004079 JAA88481.1 AK403221 BAM19640.1 KQ971311 EEZ99044.1 GANO01000277 JAB59594.1 JXUM01146283 KQ570143 KXJ68444.1 GAPW01002362 JAC11236.1 GFDL01003034 JAV32011.1 CH477363 EAT42609.1 EAT42610.1 EAT42611.1 JTDY01000154 KOB78561.1 UFQS01000039 UFQT01000039 SSW98090.1 SSX18476.1 ADMH02001074 ETN64167.1 AJWK01029436 AJWK01029437 AJWK01029438 AAAB01008933 EAA09900.4 APCN01005588 GFDF01004036 JAV10048.1 ATLV01022275 KE525331 KFB47370.1 GFDF01004035 JAV10049.1 AXCN02002042 AXCM01001028 GGFJ01006975 MBW56116.1 GGFK01008926 MBW42247.1 GBGD01002011 JAC86878.1 APGK01054399 APGK01054400 KB741249 ENN71926.1 BT126789 KB631760 AEE61751.1 ERL85899.1 GL887908 EGI69563.1 GGFK01008978 MBW42299.1 ADTU01018832 KQ981693 KYN37754.1 GGFM01004586 MBW25337.1 GFTR01005304 JAW11122.1 KQ976692 KYM77841.1 KQ436240 KOX67349.1 KZ288349 PBC27417.1 JR050887 AEY61377.1 KQ414612 KOC69144.1 KK852726 KDR17659.1 LBMM01000528 KMQ98132.1 GDKW01000330 JAI56265.1 ACPB03017890 KK107021 QOIP01000011 EZA62395.1 RLU17077.1 AK417755 BAN20970.1 NNAY01001765 OXU22977.1 GL445250 EFN89970.1 AAZX01004049 GECZ01032448 GECZ01011988 GECZ01009674 JAS37321.1 JAS57781.1 JAS60095.1 GEBQ01000403 JAT39574.1 KM221889 AIU47030.1 GEZM01058077 JAV71922.1 GL766616 EFZ14926.1 AE014134 AY121650 AAF57232.2 AAM51977.1 GEDC01002940 JAS34358.1 KQ978881 KYN27699.1 CP012523 ALC39831.1 CM000158 EDW89918.1 CH964214 EDW80345.1 CH902624 EDV33324.1 EZ423286 ADD19562.1 CM000361 CM002910 EDX05756.1 KMY91385.1 CH480843 EDW49717.1 CH954179 EDV54371.1 CH916368 EDW03548.1 GBHO01036467 GBHO01035468 GBHO01030840 GBHO01021132 GBHO01021131 GBHO01021129 GBHO01021126 GBHO01021125 GBHO01021124 GBRD01017077 GBRD01017074 GDHC01021752 GDHC01012825 JAG07137.1 JAG08136.1 JAG12764.1 JAG22472.1 JAG22473.1 JAG22475.1 JAG22478.1 JAG22479.1 JAG22480.1 JAG48750.1 JAP96876.1 JAQ05804.1 GAKP01002901 JAC56051.1 GDHF01031245 GDHF01017939 GDHF01017218 JAI21069.1 JAI34375.1 JAI35096.1 GBXI01016493 JAC97798.1 OUUW01000022 SPP89531.1 CH379058 EAL34337.2 CH479184 EDW37207.1 GFWV01005883 MAA30613.1 SPP89530.1 DS235341 EEB14998.1 AK339856 BAH70564.1 JXJN01002512 GBZX01001487 JAG91253.1 GFAC01001174 JAT98014.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000007266
+ More
UP000069940 UP000249989 UP000008820 UP000037510 UP000000673 UP000092461 UP000076407 UP000007062 UP000075840 UP000075903 UP000075882 UP000030765 UP000075881 UP000075886 UP000075900 UP000075883 UP000075880 UP000069272 UP000019118 UP000030742 UP000007755 UP000075884 UP000005205 UP000078541 UP000078540 UP000053105 UP000075920 UP000005203 UP000242457 UP000053825 UP000192223 UP000027135 UP000036403 UP000015103 UP000053097 UP000279307 UP000215335 UP000008237 UP000002358 UP000000803 UP000078492 UP000092553 UP000092445 UP000002282 UP000007798 UP000007801 UP000192221 UP000000304 UP000078200 UP000001292 UP000008711 UP000001070 UP000268350 UP000001819 UP000008744 UP000092443 UP000076408 UP000009046 UP000075885 UP000092460 UP000075902
UP000069940 UP000249989 UP000008820 UP000037510 UP000000673 UP000092461 UP000076407 UP000007062 UP000075840 UP000075903 UP000075882 UP000030765 UP000075881 UP000075886 UP000075900 UP000075883 UP000075880 UP000069272 UP000019118 UP000030742 UP000007755 UP000075884 UP000005205 UP000078541 UP000078540 UP000053105 UP000075920 UP000005203 UP000242457 UP000053825 UP000192223 UP000027135 UP000036403 UP000015103 UP000053097 UP000279307 UP000215335 UP000008237 UP000002358 UP000000803 UP000078492 UP000092553 UP000092445 UP000002282 UP000007798 UP000007801 UP000192221 UP000000304 UP000078200 UP000001292 UP000008711 UP000001070 UP000268350 UP000001819 UP000008744 UP000092443 UP000076408 UP000009046 UP000075885 UP000092460 UP000075902
Interpro
SUPFAM
SSF103111
SSF103111
Gene 3D
ProteinModelPortal
H9JP43
Q75RW2
A0A194QEB3
A0A2A4JNJ0
A0A2H1WIS9
A0A212F829
+ More
A0A0N1I5N0 S4P8R4 I4DP00 D6WCK9 U5EP77 A0A182G5K0 A0A023ERF3 A0A1Q3FWL3 Q178H4 A0A0L7LTJ4 A0A336LKJ5 W5JIF2 A0A1B0CUN5 A0A182WUU7 Q7PPS8 A0A182HFV6 A0A182V963 A0A182LPY7 A0A1L8DUC6 A0A084WAX6 A0A182K5S0 A0A1L8DUE3 A0A182QAQ2 A0A182RF43 A0A182MLW4 A0A2M4BSQ1 A0A2M4ANB2 A0A182JM66 A0A182FTR9 A0A069DS64 N6TU58 J3JU76 F4W8S2 A0A1Y9H2K6 A0A2M4ANE3 A0A158NKG6 A0A195FCI4 A0A2M3Z9X2 A0A224XRE9 A0A195B058 A0A0M8ZQ82 A0A182VT72 A0A088AJL2 A0A2A3E6P2 V9IL20 A0A0L7RE91 A0A1W4WAD1 A0A067R5R7 A0A0J7L5B0 A0A0P4VUN4 T1IC42 A0A026X249 R4WJL5 A0A232EXF5 E2B301 K7IUI9 A0A1B6EHD5 A0A1B6MUF2 A0A097PHM9 A0A1Y1LE46 E9IWZ4 Q9V9Q4 A0A1B6E8U8 A0A195EIL0 A0A0M5IWS2 A0A1A9ZR92 B4P6G1 B4N7K6 B3MTZ5 A0A1W4V286 D3TPD4 B4Q4F9 A0A1A9UWA3 B4IIG7 B3NKR7 B4JE28 A0A0A9XP10 A0A034WKD6 A0A0K8U2T3 A0A0A1WFN8 A0A3B0KVW5 Q29PF6 B4GKP1 A0A293LPH8 A0A3B0KLN3 A0A1A9YJL3 A0A182YF80 E0VNP2 A0A182PWG7 C4WRP4 A0A1B0ARP8 A0A182U942 A0A0C9SE15 A0A1E1XFF7
A0A0N1I5N0 S4P8R4 I4DP00 D6WCK9 U5EP77 A0A182G5K0 A0A023ERF3 A0A1Q3FWL3 Q178H4 A0A0L7LTJ4 A0A336LKJ5 W5JIF2 A0A1B0CUN5 A0A182WUU7 Q7PPS8 A0A182HFV6 A0A182V963 A0A182LPY7 A0A1L8DUC6 A0A084WAX6 A0A182K5S0 A0A1L8DUE3 A0A182QAQ2 A0A182RF43 A0A182MLW4 A0A2M4BSQ1 A0A2M4ANB2 A0A182JM66 A0A182FTR9 A0A069DS64 N6TU58 J3JU76 F4W8S2 A0A1Y9H2K6 A0A2M4ANE3 A0A158NKG6 A0A195FCI4 A0A2M3Z9X2 A0A224XRE9 A0A195B058 A0A0M8ZQ82 A0A182VT72 A0A088AJL2 A0A2A3E6P2 V9IL20 A0A0L7RE91 A0A1W4WAD1 A0A067R5R7 A0A0J7L5B0 A0A0P4VUN4 T1IC42 A0A026X249 R4WJL5 A0A232EXF5 E2B301 K7IUI9 A0A1B6EHD5 A0A1B6MUF2 A0A097PHM9 A0A1Y1LE46 E9IWZ4 Q9V9Q4 A0A1B6E8U8 A0A195EIL0 A0A0M5IWS2 A0A1A9ZR92 B4P6G1 B4N7K6 B3MTZ5 A0A1W4V286 D3TPD4 B4Q4F9 A0A1A9UWA3 B4IIG7 B3NKR7 B4JE28 A0A0A9XP10 A0A034WKD6 A0A0K8U2T3 A0A0A1WFN8 A0A3B0KVW5 Q29PF6 B4GKP1 A0A293LPH8 A0A3B0KLN3 A0A1A9YJL3 A0A182YF80 E0VNP2 A0A182PWG7 C4WRP4 A0A1B0ARP8 A0A182U942 A0A0C9SE15 A0A1E1XFF7
PDB
1X53
E-value=2.62055e-27,
Score=303
Ontologies
GO
PANTHER
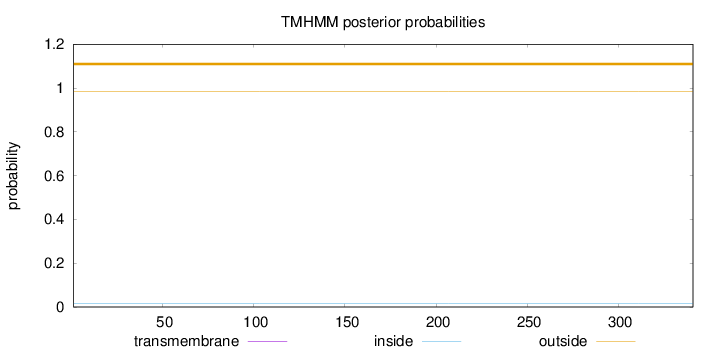
Topology
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01562
outside
1 - 341
Population Genetic Test Statistics
Pi
249.037484
Theta
190.408086
Tajima's D
1.042365
CLR
0.406091
CSRT
0.670416479176041
Interpretation
Uncertain
