Pre Gene Modal
BGIBMGA011596
Annotation
4-nitrophenylphosphatase_[Bombyx_mori]
Full name
Anoctamin
Location in the cell
Cytoplasmic Reliability : 2.518
Sequence
CDS
ATGGGTATTGAATCTAAACATTTATTAGATTTAAGTGTTGAAGACTTGCATAAATTTCTAGATTCCTTCGACCATGTCCTCTCCGATTGCGATGGTGTTATATGGACTCAAGATTCTTTGCCGCGAGTCGGAGAGTTCTTTAAACAGATGAAGAAACGCGGGAAGACAGTAAATTTCGTTTCCAATAACAGCCTTCGATCCAGAGCAAATTATGAAGCTCAATTCAAAGCAGCCAGCATCGACAACGGCTTTGAGAGCCTGATCATTCCATCTATCGCAGTGGCAGAGTACCTTAAATCAGTTACATTCAATAAAACTGTTTACTGTGTAACCTGTACTGAGACCAAAAGAGTCTTAGAAGCTCACGGATTCAAATGCAAGGAAGGTCCAGATCTAGGGCCGGAGTACTATGGCGAATACATCCAGTATTTAGAAGACGATGAAGAGATAGGAGCCGTCGTTTTTGACAGCGACTTTAAGATAAACTTGCCGAAAATGTACCGAGCCATAACCTACCTGAAAAGACCTGAGGTCCTCTTCATAAATGGGGCGACTGATAGGATGGTGCCCATGAAAACTGGTCTTTTGGGTTTAGGGACGGGAGTTTTTACAGATCTGGTTACGGTAGAAGTGAAACGAGAGCCCGTGCTGCTTGGGAAGCCAGGAAGGGTTTTCGGTGAATTCGCTATGAAGCGAGCCGGCATCACCGATCCCAGTCGAGTTCTTTTTATCGGTGACATGATCGCCCAAGATGTATCTCTAGGGAAAGCAGTTGGTTTCAACACTTTATTAGTTTTAACGAACACTACCAAAGAAGAAATGTTGTCACACACAATACGACCAGATTACTACGCGGCGTCTCTTGGTAGTATTGTGCCTCTAATTCAGTAA
Protein
MGIESKHLLDLSVEDLHKFLDSFDHVLSDCDGVIWTQDSLPRVGEFFKQMKKRGKTVNFVSNNSLRSRANYEAQFKAASIDNGFESLIIPSIAVAEYLKSVTFNKTVYCVTCTETKRVLEAHGFKCKEGPDLGPEYYGEYIQYLEDDEEIGAVVFDSDFKINLPKMYRAITYLKRPEVLFINGATDRMVPMKTGLLGLGTGVFTDLVTVEVKREPVLLGKPGRVFGEFAMKRAGITDPSRVLFIGDMIAQDVSLGKAVGFNTLLVLTNTTKEEMLSHTIRPDYYAASLGSIVPLIQ
Summary
Similarity
Belongs to the HAD-like hydrolase superfamily.
Belongs to the anoctamin family.
Belongs to the anoctamin family.
Uniprot
Q1HQD3
H9JPZ1
H9JPZ0
A0A212F4M3
A0A194PKY7
A0A2A4JCE3
+ More
A0A194QPD9 A0A3S2NEB1 A0A2H1V644 A0A0L7LHW0 B4Q1I1 B3NT22 B4IK78 Q9VYT0 B4MG65 A0A1A9V6N9 B4NDR6 B4R393 A0A1W4VTD2 D3TMD7 B4GY32 B4L874 A0A3B0K622 A0A1Q3G4H0 Q29H96 A0A1A9XGN0 Q16TW0 W8CCW9 Q16MH4 T1DGC1 A0A1I8PYG2 A0A182G7T5 A0A2C9GQG6 Q7QEP8 B4JJD8 A0A0A1WV55 A0A0A1WFS7 A0A034VPM2 T1EAV7 A0A084W0Y2 T1PGZ1 A0A182JG84 A0A1I8M0J5 A0A182YD03 A0A182TTL4 A0A182NIK9 A0A1Q3G4G5 B3N1E2 A0A182H5A4 A0A182G536 A0A182V739 A0A0L0CAW9 Q16TW1 A0A1B0DIL7 A0A2M4AAD4 B0WPC4 A0A182T4S2 A0A084WPM3 A0A182G7T6 A0A1Q3FSM7 A0A2M4CW97 A0A2M4AAX4 A0A182WKN0 B0XDV6 W5J909 A0A2M4AAP8 Q0IF21 A0A182REF9 A0A2M4A9P8 A0A2M4CYK1 A0A182GDS8 A0A2M4AA26 A0A182FGY7 Q7PIJ5 A0A1B0GI13 A0A182P5I8 A0A182I1U8 A0A182V1C6 A0A182R0U5 A0A182X735 A0A182QIE3 A0A182LYK2 A0A182K6S8 A0A182WY54 A0A1L8DZF4 A0A1B0C9C4 A0A182KPK0 U5EXS2 B0WUE2 A0A182W3Z7 A0A182JTX0 A0A0A1XSS0 A0A1L8EG67 A0A0K8V3V8 A0A1L8E9K9 A0A1L8DZA8 A0A1B6K726 B4GY31 Q0IF19 B5DMQ2
A0A194QPD9 A0A3S2NEB1 A0A2H1V644 A0A0L7LHW0 B4Q1I1 B3NT22 B4IK78 Q9VYT0 B4MG65 A0A1A9V6N9 B4NDR6 B4R393 A0A1W4VTD2 D3TMD7 B4GY32 B4L874 A0A3B0K622 A0A1Q3G4H0 Q29H96 A0A1A9XGN0 Q16TW0 W8CCW9 Q16MH4 T1DGC1 A0A1I8PYG2 A0A182G7T5 A0A2C9GQG6 Q7QEP8 B4JJD8 A0A0A1WV55 A0A0A1WFS7 A0A034VPM2 T1EAV7 A0A084W0Y2 T1PGZ1 A0A182JG84 A0A1I8M0J5 A0A182YD03 A0A182TTL4 A0A182NIK9 A0A1Q3G4G5 B3N1E2 A0A182H5A4 A0A182G536 A0A182V739 A0A0L0CAW9 Q16TW1 A0A1B0DIL7 A0A2M4AAD4 B0WPC4 A0A182T4S2 A0A084WPM3 A0A182G7T6 A0A1Q3FSM7 A0A2M4CW97 A0A2M4AAX4 A0A182WKN0 B0XDV6 W5J909 A0A2M4AAP8 Q0IF21 A0A182REF9 A0A2M4A9P8 A0A2M4CYK1 A0A182GDS8 A0A2M4AA26 A0A182FGY7 Q7PIJ5 A0A1B0GI13 A0A182P5I8 A0A182I1U8 A0A182V1C6 A0A182R0U5 A0A182X735 A0A182QIE3 A0A182LYK2 A0A182K6S8 A0A182WY54 A0A1L8DZF4 A0A1B0C9C4 A0A182KPK0 U5EXS2 B0WUE2 A0A182W3Z7 A0A182JTX0 A0A0A1XSS0 A0A1L8EG67 A0A0K8V3V8 A0A1L8E9K9 A0A1L8DZA8 A0A1B6K726 B4GY31 Q0IF19 B5DMQ2
Pubmed
19121390
22118469
26354079
26227816
17994087
17550304
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 20353571 15632085 17510324 24495485 24330624 26483478 12364791 14747013 17210077 25830018 25348373 24438588 25315136 25244985 26108605 20920257 23761445 20966253
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 20353571 15632085 17510324 24495485 24330624 26483478 12364791 14747013 17210077 25830018 25348373 24438588 25315136 25244985 26108605 20920257 23761445 20966253
EMBL
DQ443119
ABF51208.1
BABH01041419
BABH01041424
AGBW02010338
OWR48686.1
+ More
KQ459601 KPI94096.1 NWSH01002031 PCG69368.1 KQ461185 KPJ07398.1 RSAL01000075 RVE48867.1 ODYU01000633 SOQ35744.1 JTDY01001124 KOB74801.1 CM000162 EDX02470.1 CH954180 EDV45991.1 CH480851 EDW51450.1 AE014298 AY070946 AAF48108.2 AAL48568.1 CH940672 EDW57388.2 KRF77748.1 CH964239 EDW81885.1 CM000366 EDX17695.1 CCAG010005975 EZ422589 ADD18865.1 CH479196 EDW27659.1 CH933814 EDW05649.2 KRG07485.1 OUUW01000015 SPP88733.1 GFDL01000347 JAV34698.1 CH379064 EAL31862.2 CH477638 EAT37968.1 GAMC01002966 JAC03590.1 CH477861 EAT35542.1 GALA01000237 JAA94615.1 JXUM01000583 KQ560108 KXJ84480.1 APCN01004001 AAAB01008846 EAA06320.5 CH916370 EDV99690.1 GBXI01011771 JAD02521.1 GBXI01016977 JAC97314.1 GAKP01015434 JAC43518.1 GAMD01000713 JAB00878.1 ATLV01019164 KE525263 KFB43876.1 KA648072 AFP62701.1 GFDL01000361 JAV34684.1 CH902654 EDV33581.1 JXUM01026111 KQ560744 KXJ81040.1 JXUM01007166 KQ560234 KXJ83699.1 JRES01000671 KNC29382.1 EAT37967.1 AJVK01006139 GGFK01004381 MBW37702.1 DS232021 EDS32174.1 ATLV01025102 KE525369 KFB52167.1 KXJ84481.1 GFDL01004435 JAV30610.1 GGFL01005303 MBW69481.1 GGFK01004467 MBW37788.1 DS232786 EDS45663.1 ADMH02001844 ETN60927.1 GGFK01004509 MBW37830.1 CH477420 EAT41252.1 GGFK01004178 MBW37499.1 GGFL01006234 MBW70412.1 JXUM01056704 KQ561922 KXJ77152.1 GGFK01004308 MBW37629.1 AAAB01008960 EAA44124.3 EDO63772.1 AJWK01011159 AJWK01011160 APCN01000096 AXCN02001229 AXCN02000411 AXCN02000412 AXCM01007579 GFDF01002275 JAV11809.1 AJWK01002220 GANO01002404 JAB57467.1 DS232104 EDS34921.1 GBXI01000245 JAD14047.1 GFDG01001226 JAV17573.1 GDHF01019074 JAI33240.1 GFDG01003378 JAV15421.1 GFDF01002274 JAV11810.1 GECU01000703 JAT07004.1 EDW27658.1 EAT41254.1 EDY72733.2
KQ459601 KPI94096.1 NWSH01002031 PCG69368.1 KQ461185 KPJ07398.1 RSAL01000075 RVE48867.1 ODYU01000633 SOQ35744.1 JTDY01001124 KOB74801.1 CM000162 EDX02470.1 CH954180 EDV45991.1 CH480851 EDW51450.1 AE014298 AY070946 AAF48108.2 AAL48568.1 CH940672 EDW57388.2 KRF77748.1 CH964239 EDW81885.1 CM000366 EDX17695.1 CCAG010005975 EZ422589 ADD18865.1 CH479196 EDW27659.1 CH933814 EDW05649.2 KRG07485.1 OUUW01000015 SPP88733.1 GFDL01000347 JAV34698.1 CH379064 EAL31862.2 CH477638 EAT37968.1 GAMC01002966 JAC03590.1 CH477861 EAT35542.1 GALA01000237 JAA94615.1 JXUM01000583 KQ560108 KXJ84480.1 APCN01004001 AAAB01008846 EAA06320.5 CH916370 EDV99690.1 GBXI01011771 JAD02521.1 GBXI01016977 JAC97314.1 GAKP01015434 JAC43518.1 GAMD01000713 JAB00878.1 ATLV01019164 KE525263 KFB43876.1 KA648072 AFP62701.1 GFDL01000361 JAV34684.1 CH902654 EDV33581.1 JXUM01026111 KQ560744 KXJ81040.1 JXUM01007166 KQ560234 KXJ83699.1 JRES01000671 KNC29382.1 EAT37967.1 AJVK01006139 GGFK01004381 MBW37702.1 DS232021 EDS32174.1 ATLV01025102 KE525369 KFB52167.1 KXJ84481.1 GFDL01004435 JAV30610.1 GGFL01005303 MBW69481.1 GGFK01004467 MBW37788.1 DS232786 EDS45663.1 ADMH02001844 ETN60927.1 GGFK01004509 MBW37830.1 CH477420 EAT41252.1 GGFK01004178 MBW37499.1 GGFL01006234 MBW70412.1 JXUM01056704 KQ561922 KXJ77152.1 GGFK01004308 MBW37629.1 AAAB01008960 EAA44124.3 EDO63772.1 AJWK01011159 AJWK01011160 APCN01000096 AXCN02001229 AXCN02000411 AXCN02000412 AXCM01007579 GFDF01002275 JAV11809.1 AJWK01002220 GANO01002404 JAB57467.1 DS232104 EDS34921.1 GBXI01000245 JAD14047.1 GFDG01001226 JAV17573.1 GDHF01019074 JAI33240.1 GFDG01003378 JAV15421.1 GFDF01002274 JAV11810.1 GECU01000703 JAT07004.1 EDW27658.1 EAT41254.1 EDY72733.2
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000283053
+ More
UP000037510 UP000002282 UP000008711 UP000001292 UP000000803 UP000008792 UP000078200 UP000007798 UP000000304 UP000192221 UP000092444 UP000008744 UP000009192 UP000268350 UP000001819 UP000092443 UP000008820 UP000095300 UP000069940 UP000249989 UP000075840 UP000007062 UP000001070 UP000030765 UP000075880 UP000095301 UP000076408 UP000075902 UP000075884 UP000007801 UP000075903 UP000037069 UP000092462 UP000002320 UP000075901 UP000075920 UP000000673 UP000075900 UP000069272 UP000092461 UP000075885 UP000075886 UP000076407 UP000075883 UP000075881 UP000075882
UP000037510 UP000002282 UP000008711 UP000001292 UP000000803 UP000008792 UP000078200 UP000007798 UP000000304 UP000192221 UP000092444 UP000008744 UP000009192 UP000268350 UP000001819 UP000092443 UP000008820 UP000095300 UP000069940 UP000249989 UP000075840 UP000007062 UP000001070 UP000030765 UP000075880 UP000095301 UP000076408 UP000075902 UP000075884 UP000007801 UP000075903 UP000037069 UP000092462 UP000002320 UP000075901 UP000075920 UP000000673 UP000075900 UP000069272 UP000092461 UP000075885 UP000075886 UP000076407 UP000075883 UP000075881 UP000075882
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
Q1HQD3
H9JPZ1
H9JPZ0
A0A212F4M3
A0A194PKY7
A0A2A4JCE3
+ More
A0A194QPD9 A0A3S2NEB1 A0A2H1V644 A0A0L7LHW0 B4Q1I1 B3NT22 B4IK78 Q9VYT0 B4MG65 A0A1A9V6N9 B4NDR6 B4R393 A0A1W4VTD2 D3TMD7 B4GY32 B4L874 A0A3B0K622 A0A1Q3G4H0 Q29H96 A0A1A9XGN0 Q16TW0 W8CCW9 Q16MH4 T1DGC1 A0A1I8PYG2 A0A182G7T5 A0A2C9GQG6 Q7QEP8 B4JJD8 A0A0A1WV55 A0A0A1WFS7 A0A034VPM2 T1EAV7 A0A084W0Y2 T1PGZ1 A0A182JG84 A0A1I8M0J5 A0A182YD03 A0A182TTL4 A0A182NIK9 A0A1Q3G4G5 B3N1E2 A0A182H5A4 A0A182G536 A0A182V739 A0A0L0CAW9 Q16TW1 A0A1B0DIL7 A0A2M4AAD4 B0WPC4 A0A182T4S2 A0A084WPM3 A0A182G7T6 A0A1Q3FSM7 A0A2M4CW97 A0A2M4AAX4 A0A182WKN0 B0XDV6 W5J909 A0A2M4AAP8 Q0IF21 A0A182REF9 A0A2M4A9P8 A0A2M4CYK1 A0A182GDS8 A0A2M4AA26 A0A182FGY7 Q7PIJ5 A0A1B0GI13 A0A182P5I8 A0A182I1U8 A0A182V1C6 A0A182R0U5 A0A182X735 A0A182QIE3 A0A182LYK2 A0A182K6S8 A0A182WY54 A0A1L8DZF4 A0A1B0C9C4 A0A182KPK0 U5EXS2 B0WUE2 A0A182W3Z7 A0A182JTX0 A0A0A1XSS0 A0A1L8EG67 A0A0K8V3V8 A0A1L8E9K9 A0A1L8DZA8 A0A1B6K726 B4GY31 Q0IF19 B5DMQ2
A0A194QPD9 A0A3S2NEB1 A0A2H1V644 A0A0L7LHW0 B4Q1I1 B3NT22 B4IK78 Q9VYT0 B4MG65 A0A1A9V6N9 B4NDR6 B4R393 A0A1W4VTD2 D3TMD7 B4GY32 B4L874 A0A3B0K622 A0A1Q3G4H0 Q29H96 A0A1A9XGN0 Q16TW0 W8CCW9 Q16MH4 T1DGC1 A0A1I8PYG2 A0A182G7T5 A0A2C9GQG6 Q7QEP8 B4JJD8 A0A0A1WV55 A0A0A1WFS7 A0A034VPM2 T1EAV7 A0A084W0Y2 T1PGZ1 A0A182JG84 A0A1I8M0J5 A0A182YD03 A0A182TTL4 A0A182NIK9 A0A1Q3G4G5 B3N1E2 A0A182H5A4 A0A182G536 A0A182V739 A0A0L0CAW9 Q16TW1 A0A1B0DIL7 A0A2M4AAD4 B0WPC4 A0A182T4S2 A0A084WPM3 A0A182G7T6 A0A1Q3FSM7 A0A2M4CW97 A0A2M4AAX4 A0A182WKN0 B0XDV6 W5J909 A0A2M4AAP8 Q0IF21 A0A182REF9 A0A2M4A9P8 A0A2M4CYK1 A0A182GDS8 A0A2M4AA26 A0A182FGY7 Q7PIJ5 A0A1B0GI13 A0A182P5I8 A0A182I1U8 A0A182V1C6 A0A182R0U5 A0A182X735 A0A182QIE3 A0A182LYK2 A0A182K6S8 A0A182WY54 A0A1L8DZF4 A0A1B0C9C4 A0A182KPK0 U5EXS2 B0WUE2 A0A182W3Z7 A0A182JTX0 A0A0A1XSS0 A0A1L8EG67 A0A0K8V3V8 A0A1L8E9K9 A0A1L8DZA8 A0A1B6K726 B4GY31 Q0IF19 B5DMQ2
PDB
4BKM
E-value=1.01272e-15,
Score=202
Ontologies
KEGG
PANTHER
Topology
Subcellular location
Membrane
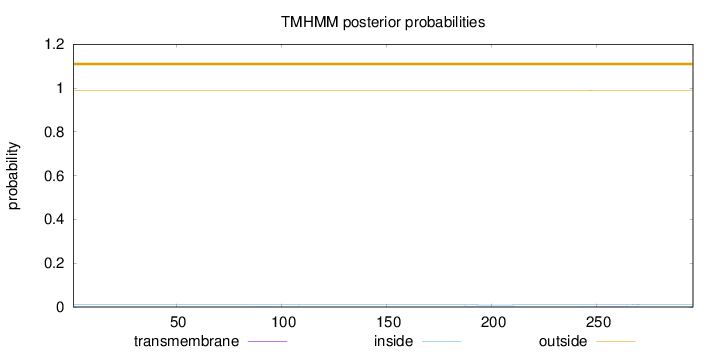
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04721
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00989
outside
1 - 296
Population Genetic Test Statistics
Pi
161.832701
Theta
167.724242
Tajima's D
-1.062709
CLR
0.783332
CSRT
0.125243737813109
Interpretation
Uncertain
