Gene
KWMTBOMO13911
Pre Gene Modal
BGIBMGA011594
Annotation
PREDICTED:_serine_proteinase_stubble_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.404 Nuclear Reliability : 1.033
Sequence
CDS
ATGCGAGCGTGCGGCGGAAACTTTCGACATAAGTACCTACGTAATTACGCGTATTTAGAACTGAAACGTTGTTACCAATTGCTATCAGGCAAGTCCTGTAAGGCGGGCAGCGCGATAGGCAAGTGCATGTTCGTGCAGGAGTGCATTAGACAAGGCGGCACCCACACTGGCGTGTGCGTCGACGGCTTCATCTTCGCGTCTTGCTGTCGGTTACCGGAGAAACCAATTAATCTAGAAGGCACCCATAATCCAGTTACAATCACGGAACGCACGCCGAATGGTGATGCGGATGCTAGTCCCCATAACAAGATTTCGTCTAGTCTTAGCATAATACCTGGGGCGAGACCCATGGCTGTTTCTGAGCAACACGCTGAAAACAGTGTTCCTGCAGCTCAGTATATGTCGAGACCAAGCAACATGAACACTGCCCATTGGCAAGTGACAACTGAGCCGAATTTCATAACGAAGCCTCGTCCATCATGGGAGAAGCCGGCGGGCAAACCGAAGCCCACAAAGAAGTTCACAACCACGACGAGTAAGCCTCACAAGAACTACTTGAAGCCGGGTTCTATGGGAACCGTTATCAACAAGATTGACCACTCAACCCCAGAAACGCAGACGACGGCCGCTACAAATGCGGCCGAGTGTGGTGTGACTTCAATGTGGCCGCGACCAGAGACCCGCATCATGGGCGGGAAGGACTCGAGCTTCGGTCGCTGGCCGTGGCAAGTGTCTGTGAGGAGGACATCTTTCTTCGGTTTTTCCTCGACCCACCGCTGCGGAGGGGCCATTATTAACGAAGGATGGATCGCAACCGCGGGGCATTGCGTTGACGACCTTCTGACCTCACAAATCAGGATTAGGGTCGGAGAATACGACTTCTCTACAGTATCAGAGGAATATCCATTTGTCGAACGTGGTGTAGCCAGAAAGGCGGTTCATCCAAAATACAATTTCTACACATACGAATACGACTTGGCGTTGGTGAAACTGGAGTCGTCCGTGCAATTCGCTCCGCATATATCGCCGATATGCTTGCCGGCCTCGGACGATTTGTTGGTTGGAGAGAACGCGACAGTCACCGGCTGGGGGAGGCTCTCGGAAGGCGGAGTACTGCCTTCGGTGTTACAAGAGGTACAAGTACCAATTGTTTCCAATGAGAGATGCAAGTCGATGTTTTTGAGGGCGGGACGACAGGAATTCATTCCTGACATTTTCCTTTGTGCGGGACACGAAAGAGGAGGCCACGATTCTTGCCAGGGAGACTCAGGCGGACCCTTACAGGTTAAAGGCAAAGATCAAAGATATTTCCTAGCTGGTATAATCAGCTGGGGAATCGGATGCGGCGAAGCAAACTTACCGGGTGTGTGTACGAGAATATCAAAATTCGTTCCCTGGATCCTACAGACTGTTAACTCATAA
Protein
MRACGGNFRHKYLRNYAYLELKRCYQLLSGKSCKAGSAIGKCMFVQECIRQGGTHTGVCVDGFIFASCCRLPEKPINLEGTHNPVTITERTPNGDADASPHNKISSSLSIIPGARPMAVSEQHAENSVPAAQYMSRPSNMNTAHWQVTTEPNFITKPRPSWEKPAGKPKPTKKFTTTTSKPHKNYLKPGSMGTVINKIDHSTPETQTTAATNAAECGVTSMWPRPETRIMGGKDSSFGRWPWQVSVRRTSFFGFSSTHRCGGAIINEGWIATAGHCVDDLLTSQIRIRVGEYDFSTVSEEYPFVERGVARKAVHPKYNFYTYEYDLALVKLESSVQFAPHISPICLPASDDLLVGENATVTGWGRLSEGGVLPSVLQEVQVPIVSNERCKSMFLRAGRQEFIPDIFLCAGHERGGHDSCQGDSGGPLQVKGKDQRYFLAGIISWGIGCGEANLPGVCTRISKFVPWILQTVNS
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01015576
BABH01015577
RSAL01000081
RVE48585.1
AGBW02014833
OWR41006.1
+ More
ODYU01000871 SOQ36284.1 KQ459601 KPI94092.1 JTDY01000849 KOB75680.1 GEDC01002043 JAS35255.1 GDHC01004671 JAQ13958.1 GDHC01012758 JAQ05871.1 GBRD01001175 JAG64646.1 GBRD01001174 JAG64647.1 GEZM01055009 GEZM01055007 JAV73254.1
ODYU01000871 SOQ36284.1 KQ459601 KPI94092.1 JTDY01000849 KOB75680.1 GEDC01002043 JAS35255.1 GDHC01004671 JAQ13958.1 GDHC01012758 JAQ05871.1 GBRD01001175 JAG64646.1 GBRD01001174 JAG64647.1 GEZM01055009 GEZM01055007 JAV73254.1
Proteomes
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
6I44
E-value=1.56014e-44,
Score=453
Ontologies
GO
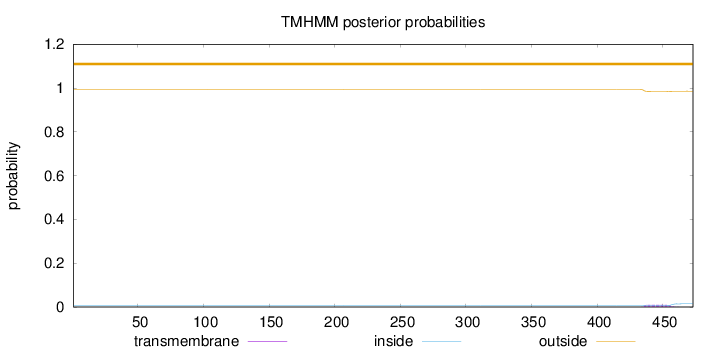
Topology
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20809
Exp number, first 60 AAs:
0.000990000000000001
Total prob of N-in:
0.00693
outside
1 - 473
Population Genetic Test Statistics
Pi
282.423139
Theta
179.578149
Tajima's D
1.620312
CLR
0.158537
CSRT
0.818359082045898
Interpretation
Uncertain
