Gene
KWMTBOMO13909
Annotation
PREDICTED:_probable_E3_SUMO-protein_ligase_RNF212_[Plutella_xylostella]
Full name
Probable E3 SUMO-protein ligase RNF212
Alternative Name
Probable E3 SUMO-protein transferase RNF212
RING finger protein 212
RING finger protein 212
Location in the cell
Extracellular Reliability : 2.899
Sequence
CDS
ATGGATTGGATTCACTGTAATAAATGTTTTTCACAATTAGAACCAGGGATAACACTGCATCTCACATCATGTGGTCACATGTTCTGCAACAATTGTTTGGGCAACGGTCTAAAAGACAGCACGTGTCTAGTATGTCGAATGCCGTGTTCTGCTATGAAATTAGTGCCAGACATGAAACAAGAAATCCAAGATTACTTCACTGATCCTGAAGAGTTGATAAAGAAGTCTTGCGATGTGATACAGTTTCAAAGGCAGCATAGAAGGAGATTGCTCTCTTATCTATTACAATCGACAGAATATTTGGCCCTTCAACATACAATATTCCTCGTTATACCATAA
Protein
MDWIHCNKCFSQLEPGITLHLTSCGHMFCNNCLGNGLKDSTCLVCRMPCSAMKLVPDMKQEIQDYFTDPEELIKKSCDVIQFQRQHRRRLLSYLLQSTEYLALQHTIFLVIP
Summary
Description
SUMO E3 ligase that acts as a regulator of crossing-over during meiosis: required to couple chromosome synapsis to the formation of crossover-specific recombination complexes. Localizes to recombination sites and stabilizes meiosis-specific recombination factors, such as MutS-gamma complex proteins (MSH4 and MSH5) and TEX11. May mediate sumoylation of target proteins MSH4 and/or MSH5, leading to enhance their binding to recombination sites. Acts as a limiting factor for crossover designation and/or reinforcement and plays an antagonist role with CCNB1IP1/HEI10 in the regulation of meiotic recombination (By similarity).
SUMO E3 ligase that acts as a regulator of crossing-over during meiosis: required to couple chromosome synapsis to the formation of crossover-specific recombination complexes. Localizes to recombination sites and stabilizes meiosis-specific recombination factors, such as MutS-gamma complex proteins (MSH4 and MSH5) and TEX11. May mediate sumoylation of target proteins MSH4 and/or MSH5, leading to enhance their binding to recombination sites. Acts as a limiting factor for crossover designation and/or reinforcement and plays an antagonist role with CCNB1IP1/HEI10 in the regulation of meiotic recombination.
SUMO E3 ligase that acts as a regulator of crossing-over during meiosis: required to couple chromosome synapsis to the formation of crossover-specific recombination complexes. Localizes to recombination sites and stabilizes meiosis-specific recombination factors, such as MutS-gamma complex proteins (MSH4 and MSH5) and TEX11. May mediate sumoylation of target proteins MSH4 and/or MSH5, leading to enhance their binding to recombination sites. Acts as a limiting factor for crossover designation and/or reinforcement and plays an antagonist role with CCNB1IP1/HEI10 in the regulation of meiotic recombination.
Keywords
Alternative splicing
Chromosome
Coiled coil
Complete proteome
Meiosis
Metal-binding
Nucleus
Polymorphism
Reference proteome
Transferase
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain Probable E3 SUMO-protein ligase RNF212
splice variant In isoform 4.
sequence variant In dbSNP:rs17728127.
splice variant In isoform 4.
sequence variant In dbSNP:rs17728127.
Uniprot
A0A2H1VDU3
A0A212FLD2
A0A3S2PDX4
A0A0L7KYW6
A0A0L8HKF0
V4ACQ1
+ More
E0VQH4 A0A2P8Z4H5 A0A210PJA5 A0A3Q7QM18 N6UIR4 A0A1B6K6J9 N6TMC8 W5MYF7 K1QA75 Q495C1-2 A0A2J8K6W5 A0A3Q0D2L6 A0A2J8SS90 A0A087TJL3 A0A1A9UZU5 A0A2U3ZBN5 G1MEN0 A0A2B4SQW2 A0A1U8C3M2 G1MEM9 A0A1X7VFA6 Q495C1-3 A0A067QNJ4 A0A2R9CIT1 A0A2I3SVD5 H2PCN2 J9P767 Q495C1-5 A0A1A9ZZQ6 A0A2J8SS60 A0A2J8K6W2 A0A2R9CLJ5 H2QP31 Q495C1 G3RU07 A0A2K5X0J2 A0A0D9RSM5 A0A3Q7UXW1 A0A1B6FPX5 A0A2I3LEY2 G5BCJ8 A0A1X7TAA9 A0A1S3WUU2 A0A287AWA2 A0A2K6CTJ7 A0A1D5QA59 A0A1X7UF31 A0A096MRK0 A0A2Y9G2V5 A0A2Y9LZH0 A0A091DSG2 A0A1B6D4S8 A0A2K5SH72 A0A2K5SH67 A0A0N8ESI7 C7BVP5 A0A2Y9JUJ1 M3YIG3 A0A3Q0DQK5 A0A1B0C2W0 A0A1A9XN25 A0A3Q2KLZ7 A0A1W3JQD8 A0A1B6DQQ5 F7APR6 A0A340Y868 A0A183CH91 A0A0G2JYP9 A0A1X7SZM7 A0A2Y9F343 A0A158P7H7 A0A091MLL2 F7GUW4 A0A0N4XXL8 F6TQD1 A0A1W5AKY9 A0A2J8SS78 A0A0P4W9L9 A0A0R3PBN3 A0A0N4U2A1 A0A2R8MHL8 D6X225 A0A091R8Q9 A0A384A2H2 A0A3P4S6I3 A0A2I2UL28 A0A2K5D1L6 A0A3R7QIU3 A0A091IEM4 A0A093EM11 A0A337RWQ1 A0A368FGJ1 A0A093P6V3
E0VQH4 A0A2P8Z4H5 A0A210PJA5 A0A3Q7QM18 N6UIR4 A0A1B6K6J9 N6TMC8 W5MYF7 K1QA75 Q495C1-2 A0A2J8K6W5 A0A3Q0D2L6 A0A2J8SS90 A0A087TJL3 A0A1A9UZU5 A0A2U3ZBN5 G1MEN0 A0A2B4SQW2 A0A1U8C3M2 G1MEM9 A0A1X7VFA6 Q495C1-3 A0A067QNJ4 A0A2R9CIT1 A0A2I3SVD5 H2PCN2 J9P767 Q495C1-5 A0A1A9ZZQ6 A0A2J8SS60 A0A2J8K6W2 A0A2R9CLJ5 H2QP31 Q495C1 G3RU07 A0A2K5X0J2 A0A0D9RSM5 A0A3Q7UXW1 A0A1B6FPX5 A0A2I3LEY2 G5BCJ8 A0A1X7TAA9 A0A1S3WUU2 A0A287AWA2 A0A2K6CTJ7 A0A1D5QA59 A0A1X7UF31 A0A096MRK0 A0A2Y9G2V5 A0A2Y9LZH0 A0A091DSG2 A0A1B6D4S8 A0A2K5SH72 A0A2K5SH67 A0A0N8ESI7 C7BVP5 A0A2Y9JUJ1 M3YIG3 A0A3Q0DQK5 A0A1B0C2W0 A0A1A9XN25 A0A3Q2KLZ7 A0A1W3JQD8 A0A1B6DQQ5 F7APR6 A0A340Y868 A0A183CH91 A0A0G2JYP9 A0A1X7SZM7 A0A2Y9F343 A0A158P7H7 A0A091MLL2 F7GUW4 A0A0N4XXL8 F6TQD1 A0A1W5AKY9 A0A2J8SS78 A0A0P4W9L9 A0A0R3PBN3 A0A0N4U2A1 A0A2R8MHL8 D6X225 A0A091R8Q9 A0A384A2H2 A0A3P4S6I3 A0A2I2UL28 A0A2K5D1L6 A0A3R7QIU3 A0A091IEM4 A0A093EM11 A0A337RWQ1 A0A368FGJ1 A0A093P6V3
EC Number
2.3.2.-
Pubmed
EMBL
ODYU01002016
SOQ38999.1
AGBW02007799
OWR54553.1
RSAL01000081
RVE48582.1
+ More
JTDY01004398 KOB68214.1 KQ417924 KOF89682.1 KB202284 ESO91101.1 DS235430 EEB15630.1 PYGN01000200 PSN51377.1 NEDP02076567 OWF36567.1 APGK01018569 APGK01018570 KB740081 ENN81615.1 GECU01000923 JAT06784.1 ENN81614.1 AHAT01001562 AHAT01001563 JH817484 EKC30858.1 AK096160 AC019103 AC092535 BC036250 BC050356 BC101258 BC101259 BC101260 BC101261 NBAG03000389 PNI30760.1 NDHI03003552 PNJ23623.1 KK115511 KFM65302.1 ACTA01067697 ACTA01075697 ACTA01083697 ACTA01091697 LSMT01000025 PFX32291.1 KK853123 KDR11067.1 AJFE02035425 AJFE02035426 AJFE02035427 AJFE02035428 AJFE02035429 AJFE02035430 AC194569 PNI30756.1 ABGA01011469 ABGA01011470 ABGA01011471 ABGA01011472 AAEX03002646 PNJ23620.1 PNI30761.1 CABD030026647 CABD030026648 CABD030026649 AQIA01053416 AQIA01053417 AQIA01053418 AQIA01053419 AQIA01053420 AQIA01053421 AQIB01147700 GECZ01017504 JAS52265.1 AHZZ02026141 JH169543 EHB07009.1 AEMK02000062 JSUE03032726 JSUE03032727 KN123337 KFO25691.1 GEDC01016713 JAS20585.1 GEBF01007067 JAN96565.1 FM207699 CAR63560.1 AEYP01079023 JXJN01024710 GEDC01009298 JAS28000.1 EAAA01002659 AABR07072361 AC117047 KK831536 KFP76862.1 UYSL01019922 VDL71338.1 AK089585 AC123743 PNJ23624.1 GDRN01076925 JAI62823.1 UYYA01000150 VDM52709.1 UYYG01001151 VDN55179.1 KQ971371 EFA10202.1 KK715081 KFQ35836.1 CYRY02046175 VCX41821.1 AANG04001605 QCYY01002627 ROT68851.1 KL218617 KFP07064.1 KK371807 KFV43944.1 JOJR01001659 RCN30109.1 KL225479 KFW72653.1
JTDY01004398 KOB68214.1 KQ417924 KOF89682.1 KB202284 ESO91101.1 DS235430 EEB15630.1 PYGN01000200 PSN51377.1 NEDP02076567 OWF36567.1 APGK01018569 APGK01018570 KB740081 ENN81615.1 GECU01000923 JAT06784.1 ENN81614.1 AHAT01001562 AHAT01001563 JH817484 EKC30858.1 AK096160 AC019103 AC092535 BC036250 BC050356 BC101258 BC101259 BC101260 BC101261 NBAG03000389 PNI30760.1 NDHI03003552 PNJ23623.1 KK115511 KFM65302.1 ACTA01067697 ACTA01075697 ACTA01083697 ACTA01091697 LSMT01000025 PFX32291.1 KK853123 KDR11067.1 AJFE02035425 AJFE02035426 AJFE02035427 AJFE02035428 AJFE02035429 AJFE02035430 AC194569 PNI30756.1 ABGA01011469 ABGA01011470 ABGA01011471 ABGA01011472 AAEX03002646 PNJ23620.1 PNI30761.1 CABD030026647 CABD030026648 CABD030026649 AQIA01053416 AQIA01053417 AQIA01053418 AQIA01053419 AQIA01053420 AQIA01053421 AQIB01147700 GECZ01017504 JAS52265.1 AHZZ02026141 JH169543 EHB07009.1 AEMK02000062 JSUE03032726 JSUE03032727 KN123337 KFO25691.1 GEDC01016713 JAS20585.1 GEBF01007067 JAN96565.1 FM207699 CAR63560.1 AEYP01079023 JXJN01024710 GEDC01009298 JAS28000.1 EAAA01002659 AABR07072361 AC117047 KK831536 KFP76862.1 UYSL01019922 VDL71338.1 AK089585 AC123743 PNJ23624.1 GDRN01076925 JAI62823.1 UYYA01000150 VDM52709.1 UYYG01001151 VDN55179.1 KQ971371 EFA10202.1 KK715081 KFQ35836.1 CYRY02046175 VCX41821.1 AANG04001605 QCYY01002627 ROT68851.1 KL218617 KFP07064.1 KK371807 KFV43944.1 JOJR01001659 RCN30109.1 KL225479 KFW72653.1
Proteomes
UP000007151
UP000283053
UP000037510
UP000053454
UP000030746
UP000009046
+ More
UP000245037 UP000242188 UP000286641 UP000019118 UP000018468 UP000005408 UP000005640 UP000189706 UP000054359 UP000078200 UP000245340 UP000008912 UP000225706 UP000007879 UP000027135 UP000240080 UP000002277 UP000001595 UP000002254 UP000092445 UP000001519 UP000233100 UP000029965 UP000286642 UP000028761 UP000006813 UP000079721 UP000008227 UP000233120 UP000006718 UP000248481 UP000248483 UP000028990 UP000233040 UP000248482 UP000000715 UP000189704 UP000092460 UP000092443 UP000002281 UP000008144 UP000265300 UP000050741 UP000002494 UP000035642 UP000008225 UP000038043 UP000271162 UP000000589 UP000192224 UP000050601 UP000267027 UP000038040 UP000274756 UP000007266 UP000261681 UP000011712 UP000233020 UP000283509 UP000054308 UP000252519 UP000054081
UP000245037 UP000242188 UP000286641 UP000019118 UP000018468 UP000005408 UP000005640 UP000189706 UP000054359 UP000078200 UP000245340 UP000008912 UP000225706 UP000007879 UP000027135 UP000240080 UP000002277 UP000001595 UP000002254 UP000092445 UP000001519 UP000233100 UP000029965 UP000286642 UP000028761 UP000006813 UP000079721 UP000008227 UP000233120 UP000006718 UP000248481 UP000248483 UP000028990 UP000233040 UP000248482 UP000000715 UP000189704 UP000092460 UP000092443 UP000002281 UP000008144 UP000265300 UP000050741 UP000002494 UP000035642 UP000008225 UP000038043 UP000271162 UP000000589 UP000192224 UP000050601 UP000267027 UP000038040 UP000274756 UP000007266 UP000261681 UP000011712 UP000233020 UP000283509 UP000054308 UP000252519 UP000054081
Pfam
PF14634 zf-RING_5
Interpro
SUPFAM
SSF89837
SSF89837
Gene 3D
ProteinModelPortal
A0A2H1VDU3
A0A212FLD2
A0A3S2PDX4
A0A0L7KYW6
A0A0L8HKF0
V4ACQ1
+ More
E0VQH4 A0A2P8Z4H5 A0A210PJA5 A0A3Q7QM18 N6UIR4 A0A1B6K6J9 N6TMC8 W5MYF7 K1QA75 Q495C1-2 A0A2J8K6W5 A0A3Q0D2L6 A0A2J8SS90 A0A087TJL3 A0A1A9UZU5 A0A2U3ZBN5 G1MEN0 A0A2B4SQW2 A0A1U8C3M2 G1MEM9 A0A1X7VFA6 Q495C1-3 A0A067QNJ4 A0A2R9CIT1 A0A2I3SVD5 H2PCN2 J9P767 Q495C1-5 A0A1A9ZZQ6 A0A2J8SS60 A0A2J8K6W2 A0A2R9CLJ5 H2QP31 Q495C1 G3RU07 A0A2K5X0J2 A0A0D9RSM5 A0A3Q7UXW1 A0A1B6FPX5 A0A2I3LEY2 G5BCJ8 A0A1X7TAA9 A0A1S3WUU2 A0A287AWA2 A0A2K6CTJ7 A0A1D5QA59 A0A1X7UF31 A0A096MRK0 A0A2Y9G2V5 A0A2Y9LZH0 A0A091DSG2 A0A1B6D4S8 A0A2K5SH72 A0A2K5SH67 A0A0N8ESI7 C7BVP5 A0A2Y9JUJ1 M3YIG3 A0A3Q0DQK5 A0A1B0C2W0 A0A1A9XN25 A0A3Q2KLZ7 A0A1W3JQD8 A0A1B6DQQ5 F7APR6 A0A340Y868 A0A183CH91 A0A0G2JYP9 A0A1X7SZM7 A0A2Y9F343 A0A158P7H7 A0A091MLL2 F7GUW4 A0A0N4XXL8 F6TQD1 A0A1W5AKY9 A0A2J8SS78 A0A0P4W9L9 A0A0R3PBN3 A0A0N4U2A1 A0A2R8MHL8 D6X225 A0A091R8Q9 A0A384A2H2 A0A3P4S6I3 A0A2I2UL28 A0A2K5D1L6 A0A3R7QIU3 A0A091IEM4 A0A093EM11 A0A337RWQ1 A0A368FGJ1 A0A093P6V3
E0VQH4 A0A2P8Z4H5 A0A210PJA5 A0A3Q7QM18 N6UIR4 A0A1B6K6J9 N6TMC8 W5MYF7 K1QA75 Q495C1-2 A0A2J8K6W5 A0A3Q0D2L6 A0A2J8SS90 A0A087TJL3 A0A1A9UZU5 A0A2U3ZBN5 G1MEN0 A0A2B4SQW2 A0A1U8C3M2 G1MEM9 A0A1X7VFA6 Q495C1-3 A0A067QNJ4 A0A2R9CIT1 A0A2I3SVD5 H2PCN2 J9P767 Q495C1-5 A0A1A9ZZQ6 A0A2J8SS60 A0A2J8K6W2 A0A2R9CLJ5 H2QP31 Q495C1 G3RU07 A0A2K5X0J2 A0A0D9RSM5 A0A3Q7UXW1 A0A1B6FPX5 A0A2I3LEY2 G5BCJ8 A0A1X7TAA9 A0A1S3WUU2 A0A287AWA2 A0A2K6CTJ7 A0A1D5QA59 A0A1X7UF31 A0A096MRK0 A0A2Y9G2V5 A0A2Y9LZH0 A0A091DSG2 A0A1B6D4S8 A0A2K5SH72 A0A2K5SH67 A0A0N8ESI7 C7BVP5 A0A2Y9JUJ1 M3YIG3 A0A3Q0DQK5 A0A1B0C2W0 A0A1A9XN25 A0A3Q2KLZ7 A0A1W3JQD8 A0A1B6DQQ5 F7APR6 A0A340Y868 A0A183CH91 A0A0G2JYP9 A0A1X7SZM7 A0A2Y9F343 A0A158P7H7 A0A091MLL2 F7GUW4 A0A0N4XXL8 F6TQD1 A0A1W5AKY9 A0A2J8SS78 A0A0P4W9L9 A0A0R3PBN3 A0A0N4U2A1 A0A2R8MHL8 D6X225 A0A091R8Q9 A0A384A2H2 A0A3P4S6I3 A0A2I2UL28 A0A2K5D1L6 A0A3R7QIU3 A0A091IEM4 A0A093EM11 A0A337RWQ1 A0A368FGJ1 A0A093P6V3
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Associates to the synaptonemal complex. Localizes to a minority of double-strand breaks (DSBs) sites. Marks crossover sites during midpachynema (By similarity). With evidence from 1 publications.
Chromosome Associates to the synaptonemal complex. Localizes to a minority of double-strand breaks (DSBs) sites. Marks crossover sites during midpachynema (By similarity). With evidence from 1 publications.
Chromosome Associates to the synaptonemal complex. Localizes to a minority of double-strand breaks (DSBs) sites. Marks crossover sites during midpachynema (By similarity). With evidence from 1 publications.
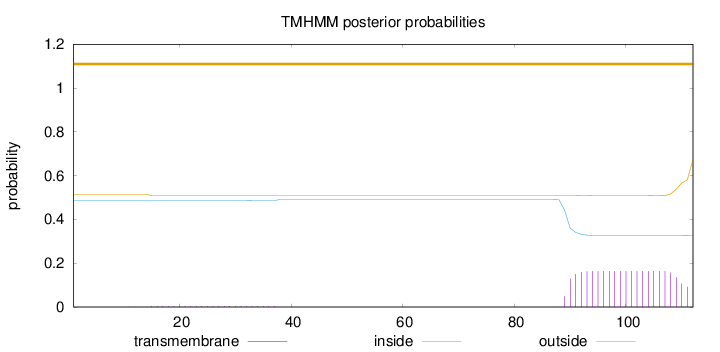
Length:
112
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.54936
Exp number, first 60 AAs:
0.10563
Total prob of N-in:
0.48599
outside
1 - 112
Population Genetic Test Statistics
Pi
187.255451
Theta
127.716559
Tajima's D
1.40331
CLR
0.138179
CSRT
0.758462076896155
Interpretation
Uncertain
