Gene
KWMTBOMO13908 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011424
Annotation
transferrin_precursor_[Bombyx_mori]
Full name
Transferrin
Location in the cell
Extracellular Reliability : 3.61
Sequence
CDS
ATGGCTCTAAAATATTTCATTTTGATAACCCTGATTTGTGCATGTGTGAACGCAGCGAAAACTACATATAAAATATGCGTGCCATCACAACACCTGAAAGCGTGTCAGGACATGGTGGATATTCCGACCAAATCCAAGGTCACACTAGACTGCATTCCCGCCAGAGATAGAATGGAGTGTCTGAATTACGTTCAGCAGAGGCAAGCCGATTTCGTTCCAGTTGACCCCGAAGACATGTACGTGGCCGCCAAAATACCCAATCAGGACTTCGTCGTTTTCCAGGAGTACAGAACCGATGAAGAACCGGATGCGCCTTTCCGTTACGAAGCAGTAATTGTCATCCATAAGGATTTACCGATAAACAATCTGGACCAGCTGAAAGGCTTGAAGTCCTGTCATACTGGAGTTAACAGAAACGTGGGATACAAGATCCCTCTAACAATGCTGATGAAGCGTGCAGTTTTCCCAAAAATGAACGACCACAGCATTTCGCCGAAGGAAAACGAACTAAAAGCCTTGTCGACGTTCTTCACGAAATCTTGCATCGTAGGTAAATGGTCTCCGGATCCGAAAACAAACTCGGCTTGGAAGGCCCAATACAACAAGCTATGCTCTATGTGCGAACATCCAGAGAGATGCGACTATCCCGACGAATTCAGCGGCTACGTGGGCGCATTGAAGTGTCTCGCTCACAACAACGGACAAGTCGCCTTCACCAAAGTCATATTCACCAGGAAATTCTTCGGATTGCCCGTCGGTACCACTCCAGCGAGTCCTTCAAACGAGAACCCTGATGAATACAGGTATCTCTGCGTGGACGGTTCTAAAGTGCCGATAAGGGACAAGGCCTGCTCTTGGGCAGCTAGACCGTGGCAGGGACTGATCGGACATAACGACGTTCTGGCTAAACTTAGCCCGCTGAGGGAGAAAATAAAGCAGTTGGCTGATGCCGGTTCCTCAAACCAACCCGAGTGGTTCACCAAAGTTCTAGGGCTATCAGACAAAATCCACTACGTGGCCGACAATATTCCGATCAAAGCCATTGACTATTTGAACAAGGCCAACTACACCGAAGTCATTGAAAGAGGTCACGGTGCTCCCGAATTAGTCGTCAGATTGTGTGTCACATCAAACGTGGCTTTAGCCAAATGCAGAGCTATGTCCGTGTTCGCGTTCAGTAGAGACATCAGACCCATCCTGGATTGTGTTCAAGAGGCTTCAGAAACCGACTGCCTTAAAAGCGTTCAAGACAATGGCTCCGATTTGGCATCAGTCGACGACATGAGGGTAGCTGCAGCAGCAAAGAAATACGTCTTGCACCCGGTCTTCCACGAGGTATACGGAGAAAAGAAAACTCCTAACTACGCTGTGGCTGTCGTCAAAAAAGGAACATCCTTCAATAAGATGGAAGATTTAAGAGGAAAGAAATCTTGTCACAGTTCGTACGGCACTTTCAGTGGACTTGACGCTCCATTGTATTATCTCATAAACAAGAGGATCATAAAACCTGATCAATGCATTAAGAACTTTGGTGATTTCTTCTCCGGCGGTTCATGTTTGCCAGGCGTCGACAAACCAGAGAACAATCCCAGCGGTGATGATGTATCATCATTAAAGAAACAATGCGGAAGTGACAGCAGTCCTTGGAAATGTCTTCAGGAAGATCGTGGCGATGTCGCTTTTGTGTCGAGTGCTGATTTATCAAACTTTGATGCTTCACAATATGAGTTGCTCTGTGTGAACCGAGAGACGGGAGGCCGTGACACTCTGTCCAACTATCCTACTTGTAACATAGCGATGGCTCCATCCAGAACCTGGCTATCAGCCAAGGACTTTTTATCAGATGTCTCTATTGCCCACACTCCACTCAGCTTAGCACAGTTATTGGACACCAGAACTGACCTGTTCAACATTTACGGAGAATTCTTGAAGAACAACAATGTCATTTTCAATAATGCTGCCAAAGGATTAGCCACAGTTGAAAATCTTGACTTTGAGAAATTCAAGTCAATCCATGATGTTATTTCTTCATGTGGCATAGCTTAA
Protein
MALKYFILITLICACVNAAKTTYKICVPSQHLKACQDMVDIPTKSKVTLDCIPARDRMECLNYVQQRQADFVPVDPEDMYVAAKIPNQDFVVFQEYRTDEEPDAPFRYEAVIVIHKDLPINNLDQLKGLKSCHTGVNRNVGYKIPLTMLMKRAVFPKMNDHSISPKENELKALSTFFTKSCIVGKWSPDPKTNSAWKAQYNKLCSMCEHPERCDYPDEFSGYVGALKCLAHNNGQVAFTKVIFTRKFFGLPVGTTPASPSNENPDEYRYLCVDGSKVPIRDKACSWAARPWQGLIGHNDVLAKLSPLREKIKQLADAGSSNQPEWFTKVLGLSDKIHYVADNIPIKAIDYLNKANYTEVIERGHGAPELVVRLCVTSNVALAKCRAMSVFAFSRDIRPILDCVQEASETDCLKSVQDNGSDLASVDDMRVAAAAKKYVLHPVFHEVYGEKKTPNYAVAVVKKGTSFNKMEDLRGKKSCHSSYGTFSGLDAPLYYLINKRIIKPDQCIKNFGDFFSGGSCLPGVDKPENNPSGDDVSSLKKQCGSDSSPWKCLQEDRGDVAFVSSADLSNFDASQYELLCVNRETGGRDTLSNYPTCNIAMAPSRTWLSAKDFLSDVSIAHTPLSLAQLLDTRTDLFNIYGEFLKNNNVIFNNAAKGLATVENLDFEKFKSIHDVISSCGIA
Summary
Description
Transferrins are iron binding transport proteins which bind Fe(3+) ion in association with the binding of an anion, usually bicarbonate.
Similarity
Belongs to the transferrin family.
Keywords
Direct protein sequencing
Disulfide bond
Glycoprotein
Ion transport
Iron
Iron transport
Metal-binding
Repeat
Secreted
Signal
Transport
Feature
chain Transferrin
Uniprot
O97158
H9JPG9
P22297
A7IT76
A0A194RH67
A0A194PSJ1
+ More
A0A212FLE3 A0JCK0 Q6F4J2 S4NYG0 A0A0B5H6A8 Q6UQ29 Q6Q2Z2 D5M9Y5 A0A2A4IY79 I4DNC0 A0A1W4WDU6 Q8MU80 Q02942 A0A067R8G2 A0A1V1FYW4 A0A2H4N2F8 A0A172WCD8 A0A2J7RCH3 H2F490 Q5FX34 Q0GB80 A0A395LHF4 A0A139WAX1 A0A1Y1MQU6 A0A1B6IXU6 A0A0L7KTB1 Q6USR2 A0A2Z6FI57 A0A0E4AVN5 A8D919 A0A0M9A935 A0A2K8JLE0 V9IAX8 Q86PH6 A0A088AFH7 A0A1B6CDF0 A0A1B6DSL2 A0A0J7KZU0 J7F1T1 A0A0L7QKR3 A0A2J7RCG2 F4W957 A0A195EDU2 A0A026WB04 A0A195B5L1 A0A158P0A5 A0A151WU63 A0A2A3ECB1 A0A151ID51 A0A146LNB5 E9I8L1 A0A0K8TJC0 A0A0A9WNT8 A0A224XL75 Q3MJL5 A0A195F369 O96418 E2B326 A0A170Y4Q2 M4WMH6 A0A3Q0IQ04 A0A1Q1NPJ1 A0A154PIR0 R4WJB4 B8LJ43 A0A310S6K8 R4G8W6 K7J4P3 A0A0C9RQZ4 A0A2Z5BV72 A0A0C9RMV8 A0A1B6J4K1 A0A146LKV4 A0A1L8DJA2 A0A1J1IUB7 Q16894 A0A3G5BIM0
A0A212FLE3 A0JCK0 Q6F4J2 S4NYG0 A0A0B5H6A8 Q6UQ29 Q6Q2Z2 D5M9Y5 A0A2A4IY79 I4DNC0 A0A1W4WDU6 Q8MU80 Q02942 A0A067R8G2 A0A1V1FYW4 A0A2H4N2F8 A0A172WCD8 A0A2J7RCH3 H2F490 Q5FX34 Q0GB80 A0A395LHF4 A0A139WAX1 A0A1Y1MQU6 A0A1B6IXU6 A0A0L7KTB1 Q6USR2 A0A2Z6FI57 A0A0E4AVN5 A8D919 A0A0M9A935 A0A2K8JLE0 V9IAX8 Q86PH6 A0A088AFH7 A0A1B6CDF0 A0A1B6DSL2 A0A0J7KZU0 J7F1T1 A0A0L7QKR3 A0A2J7RCG2 F4W957 A0A195EDU2 A0A026WB04 A0A195B5L1 A0A158P0A5 A0A151WU63 A0A2A3ECB1 A0A151ID51 A0A146LNB5 E9I8L1 A0A0K8TJC0 A0A0A9WNT8 A0A224XL75 Q3MJL5 A0A195F369 O96418 E2B326 A0A170Y4Q2 M4WMH6 A0A3Q0IQ04 A0A1Q1NPJ1 A0A154PIR0 R4WJB4 B8LJ43 A0A310S6K8 R4G8W6 K7J4P3 A0A0C9RQZ4 A0A2Z5BV72 A0A0C9RMV8 A0A1B6J4K1 A0A146LKV4 A0A1L8DJA2 A0A1J1IUB7 Q16894 A0A3G5BIM0
Pubmed
19467259
19121390
2254322
26354079
22118469
23622113
+ More
25430818 15110871 22229520 22651552 12542630 7679500 24845553 28410430 16962940 18362917 19820115 28004739 26227816 18824242 11864369 15110862 21719571 24508170 30249741 21347285 26823975 21282665 25401762 16039806 10790182 20798317 23442387 28130397 23691247 20075255 29272461 7957753 9356450 30400621
25430818 15110871 22229520 22651552 12542630 7679500 24845553 28410430 16962940 18362917 19820115 28004739 26227816 18824242 11864369 15110862 21719571 24508170 30249741 21347285 26823975 21282665 25401762 16039806 10790182 20798317 23442387 28130397 23691247 20075255 29272461 7957753 9356450 30400621
EMBL
AF119098
DQ375762
AAD18032.2
ABD35289.1
BABH01015561
BABH01015562
+ More
M62802 DQ470073 ABF21123.1 KQ460205 KPJ17168.1 KQ459601 KPI94090.1 AGBW02007799 OWR54552.1 AB282638 BAF36818.1 AB158473 BAD27263.1 GAIX01013820 JAA78740.1 KJ641611 AJF35009.1 AY364430 AAQ63970.2 AY563106 AAT08022.1 HM026347 ADF35768.1 NWSH01005123 PCG64426.1 AK402830 BAM19410.1 AF535146 AAN03488.1 L05340 KK852636 KDR19744.1 FX985456 BAX07469.1 KY653081 ATU47275.1 KU213914 ANF07122.1 NEVH01005885 PNF38525.1 JQ257002 AEX92027.1 AY894342 DQ386641 AAW70172.1 DQ882181 ABI31834.1 QRBB01000009 RDS75647.1 KQ971380 KYB25049.1 GEZM01024024 JAV88052.1 GECU01015967 JAS91739.1 JTDY01006095 KOB66291.1 AY362358 AAQ62963.2 LC384923 BBE27867.1 LC009513 BAQ94504.1 EU144233 ABV68876.1 KQ435710 KOX79720.1 MF683360 ATU82501.1 JR036928 JR036929 JR036930 JR036931 JR036932 JR036933 AEY57479.1 AEY57480.1 AEY57481.1 AEY57482.1 AEY57483.1 AEY57484.1 AY217097 AY336528 AY336529 AAO39761.1 AAQ02339.1 AAQ02340.1 GEDC01025826 JAS11472.1 GEDC01008637 JAS28661.1 LBMM01001777 KMQ95759.1 HQ828073 JF330111 AET36535.1 AET85185.1 KQ414940 KOC59155.1 PNF38524.1 GL888002 EGI69239.1 KQ979039 KYN23276.1 KK107293 QOIP01000005 EZA53240.1 RLU22580.1 KQ976598 KYM79560.1 ADTU01000981 KQ982736 KYQ51459.1 KZ288287 PBC29375.1 KQ977989 KYM98331.1 GDHC01009954 JAQ08675.1 GL761587 EFZ23092.1 GBRD01000119 JAG65702.1 GBHO01033507 JAG10097.1 GFTR01007645 JAW08781.1 AY940115 AY940116 AAY21643.1 AAY21644.1 KQ981855 KYN34908.1 AF046126 AAD02419.1 GL445255 EFN89912.1 GEMB01003665 JAR99553.1 JX560434 AGI17578.1 KX459692 AQM58443.1 KQ434902 KZC11090.1 AK417595 BAN20810.1 ACPB03016283 EF634463 ABU96701.1 KQ780658 OAD51936.1 GAHY01000257 JAA77253.1 AAZX01010569 GBYB01009696 JAG79463.1 MF627744 AXA98484.1 GBYB01009695 JAG79462.1 GECU01013606 JAS94100.1 GDHC01009926 JAQ08703.1 GFDF01007644 JAV06440.1 CVRI01000059 CRL03704.1 AF019117 AAB87414.1 MK075223 AYV99626.1
M62802 DQ470073 ABF21123.1 KQ460205 KPJ17168.1 KQ459601 KPI94090.1 AGBW02007799 OWR54552.1 AB282638 BAF36818.1 AB158473 BAD27263.1 GAIX01013820 JAA78740.1 KJ641611 AJF35009.1 AY364430 AAQ63970.2 AY563106 AAT08022.1 HM026347 ADF35768.1 NWSH01005123 PCG64426.1 AK402830 BAM19410.1 AF535146 AAN03488.1 L05340 KK852636 KDR19744.1 FX985456 BAX07469.1 KY653081 ATU47275.1 KU213914 ANF07122.1 NEVH01005885 PNF38525.1 JQ257002 AEX92027.1 AY894342 DQ386641 AAW70172.1 DQ882181 ABI31834.1 QRBB01000009 RDS75647.1 KQ971380 KYB25049.1 GEZM01024024 JAV88052.1 GECU01015967 JAS91739.1 JTDY01006095 KOB66291.1 AY362358 AAQ62963.2 LC384923 BBE27867.1 LC009513 BAQ94504.1 EU144233 ABV68876.1 KQ435710 KOX79720.1 MF683360 ATU82501.1 JR036928 JR036929 JR036930 JR036931 JR036932 JR036933 AEY57479.1 AEY57480.1 AEY57481.1 AEY57482.1 AEY57483.1 AEY57484.1 AY217097 AY336528 AY336529 AAO39761.1 AAQ02339.1 AAQ02340.1 GEDC01025826 JAS11472.1 GEDC01008637 JAS28661.1 LBMM01001777 KMQ95759.1 HQ828073 JF330111 AET36535.1 AET85185.1 KQ414940 KOC59155.1 PNF38524.1 GL888002 EGI69239.1 KQ979039 KYN23276.1 KK107293 QOIP01000005 EZA53240.1 RLU22580.1 KQ976598 KYM79560.1 ADTU01000981 KQ982736 KYQ51459.1 KZ288287 PBC29375.1 KQ977989 KYM98331.1 GDHC01009954 JAQ08675.1 GL761587 EFZ23092.1 GBRD01000119 JAG65702.1 GBHO01033507 JAG10097.1 GFTR01007645 JAW08781.1 AY940115 AY940116 AAY21643.1 AAY21644.1 KQ981855 KYN34908.1 AF046126 AAD02419.1 GL445255 EFN89912.1 GEMB01003665 JAR99553.1 JX560434 AGI17578.1 KX459692 AQM58443.1 KQ434902 KZC11090.1 AK417595 BAN20810.1 ACPB03016283 EF634463 ABU96701.1 KQ780658 OAD51936.1 GAHY01000257 JAA77253.1 AAZX01010569 GBYB01009696 JAG79463.1 MF627744 AXA98484.1 GBYB01009695 JAG79462.1 GECU01013606 JAS94100.1 GDHC01009926 JAQ08703.1 GFDF01007644 JAV06440.1 CVRI01000059 CRL03704.1 AF019117 AAB87414.1 MK075223 AYV99626.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000192223
+ More
UP000027135 UP000235965 UP000254101 UP000007266 UP000037510 UP000053105 UP000005203 UP000036403 UP000053825 UP000007755 UP000078492 UP000053097 UP000279307 UP000078540 UP000005205 UP000075809 UP000242457 UP000078542 UP000078541 UP000008237 UP000079169 UP000076502 UP000015103 UP000002358 UP000183832
UP000027135 UP000235965 UP000254101 UP000007266 UP000037510 UP000053105 UP000005203 UP000036403 UP000053825 UP000007755 UP000078492 UP000053097 UP000279307 UP000078540 UP000005205 UP000075809 UP000242457 UP000078542 UP000078541 UP000008237 UP000079169 UP000076502 UP000015103 UP000002358 UP000183832
Pfam
PF00405 Transferrin
ProteinModelPortal
O97158
H9JPG9
P22297
A7IT76
A0A194RH67
A0A194PSJ1
+ More
A0A212FLE3 A0JCK0 Q6F4J2 S4NYG0 A0A0B5H6A8 Q6UQ29 Q6Q2Z2 D5M9Y5 A0A2A4IY79 I4DNC0 A0A1W4WDU6 Q8MU80 Q02942 A0A067R8G2 A0A1V1FYW4 A0A2H4N2F8 A0A172WCD8 A0A2J7RCH3 H2F490 Q5FX34 Q0GB80 A0A395LHF4 A0A139WAX1 A0A1Y1MQU6 A0A1B6IXU6 A0A0L7KTB1 Q6USR2 A0A2Z6FI57 A0A0E4AVN5 A8D919 A0A0M9A935 A0A2K8JLE0 V9IAX8 Q86PH6 A0A088AFH7 A0A1B6CDF0 A0A1B6DSL2 A0A0J7KZU0 J7F1T1 A0A0L7QKR3 A0A2J7RCG2 F4W957 A0A195EDU2 A0A026WB04 A0A195B5L1 A0A158P0A5 A0A151WU63 A0A2A3ECB1 A0A151ID51 A0A146LNB5 E9I8L1 A0A0K8TJC0 A0A0A9WNT8 A0A224XL75 Q3MJL5 A0A195F369 O96418 E2B326 A0A170Y4Q2 M4WMH6 A0A3Q0IQ04 A0A1Q1NPJ1 A0A154PIR0 R4WJB4 B8LJ43 A0A310S6K8 R4G8W6 K7J4P3 A0A0C9RQZ4 A0A2Z5BV72 A0A0C9RMV8 A0A1B6J4K1 A0A146LKV4 A0A1L8DJA2 A0A1J1IUB7 Q16894 A0A3G5BIM0
A0A212FLE3 A0JCK0 Q6F4J2 S4NYG0 A0A0B5H6A8 Q6UQ29 Q6Q2Z2 D5M9Y5 A0A2A4IY79 I4DNC0 A0A1W4WDU6 Q8MU80 Q02942 A0A067R8G2 A0A1V1FYW4 A0A2H4N2F8 A0A172WCD8 A0A2J7RCH3 H2F490 Q5FX34 Q0GB80 A0A395LHF4 A0A139WAX1 A0A1Y1MQU6 A0A1B6IXU6 A0A0L7KTB1 Q6USR2 A0A2Z6FI57 A0A0E4AVN5 A8D919 A0A0M9A935 A0A2K8JLE0 V9IAX8 Q86PH6 A0A088AFH7 A0A1B6CDF0 A0A1B6DSL2 A0A0J7KZU0 J7F1T1 A0A0L7QKR3 A0A2J7RCG2 F4W957 A0A195EDU2 A0A026WB04 A0A195B5L1 A0A158P0A5 A0A151WU63 A0A2A3ECB1 A0A151ID51 A0A146LNB5 E9I8L1 A0A0K8TJC0 A0A0A9WNT8 A0A224XL75 Q3MJL5 A0A195F369 O96418 E2B326 A0A170Y4Q2 M4WMH6 A0A3Q0IQ04 A0A1Q1NPJ1 A0A154PIR0 R4WJB4 B8LJ43 A0A310S6K8 R4G8W6 K7J4P3 A0A0C9RQZ4 A0A2Z5BV72 A0A0C9RMV8 A0A1B6J4K1 A0A146LKV4 A0A1L8DJA2 A0A1J1IUB7 Q16894 A0A3G5BIM0
PDB
6D05
E-value=7.32964e-40,
Score=414
Ontologies
GO
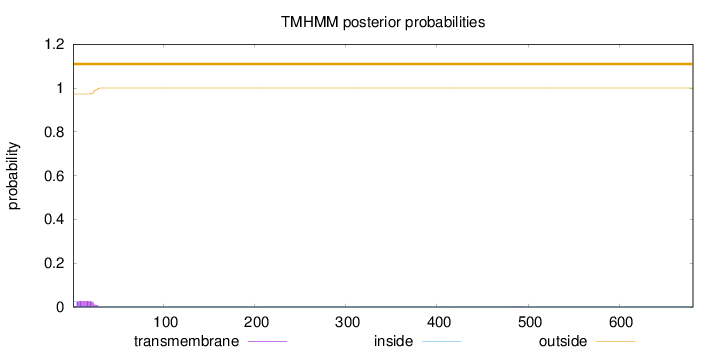
Topology
Subcellular location
Secreted
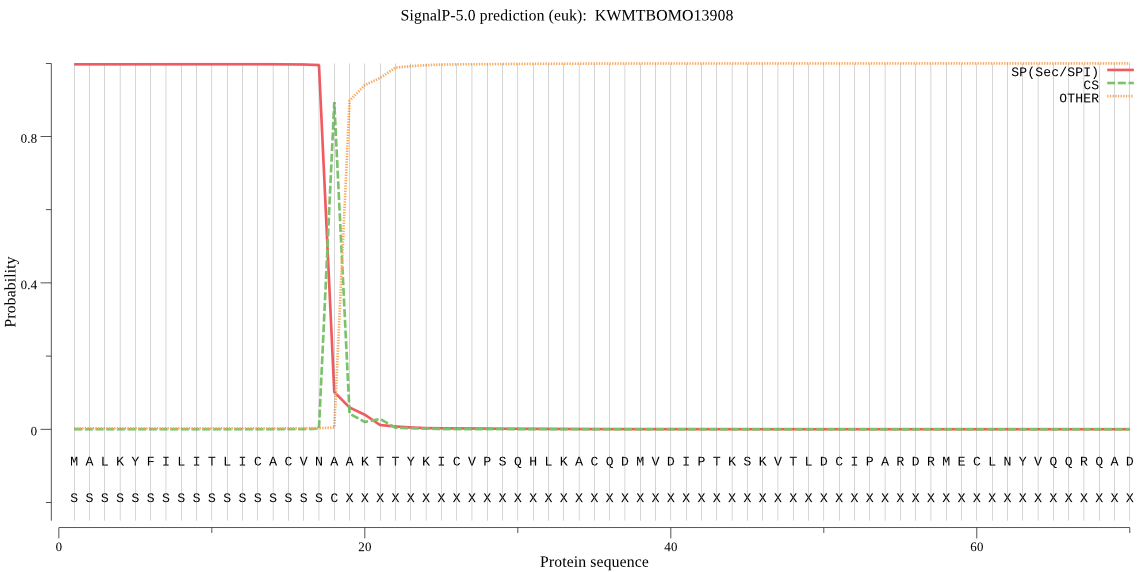
SignalP
Position: 1 - 18,
Likelihood: 0.997399
Length:
681
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.545289999999996
Exp number, first 60 AAs:
0.54221
Total prob of N-in:
0.02691
outside
1 - 681
Population Genetic Test Statistics
Pi
227.311911
Theta
193.269268
Tajima's D
0.263317
CLR
0.956434
CSRT
0.44312784360782
Interpretation
Uncertain
