Gene
KWMTBOMO13905
Pre Gene Modal
BGIBMGA011425
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_BTB/POZ_domain-containing_protein_KCTD16_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.929 Mitochondrial Reliability : 1.396
Sequence
CDS
ATGGGCTCGCGCGGCGCCGAGACGTGCGGTAAGTGGGTCAGGGAGGGAGCGGGGCGCGCAGCCGCTGCCACACTGCGCCCTCAGCCTCTAACCATGGCGGACGCTCCGCCTGAGGTCCTCGAGCTCAACGTGGGCGGCGTGCACTATGCCACTACGCGCGATACGCTGCTACGCGAGCCAGACTCGCTGCCCGCCGCCGCGCTTCAAGATCCCGACCATCCGCGCGACGCTCGTGGTCGCATCTTCTTCGATAGAGACGGCGTTCTCTTCCGCTATGTGCTCGACTATCTCAGAGACGGTGGACTCGTTTTGCCGGAGTGCTTCAGAGAACACAAGCGCCTCGCGAGGGAAGCCAAACATTACAGACTAGTGGGGCTCGAGAATGCAGTCCGAGAAGCAGATCCTCGTCCGCCAAATCGCGAGCGAGGATGCATCACGGTGGGATACCGGGGCAGTTTTGCATTCGGACGCGACGGCCTCGCCGATGTAAAGTTCCGTAAGTTGTCTCGCATTCTCGTGTGCGGTAAAGTAACGCTGTGTCGAGACGTCTTCGGCGAGACATTGAACGAGTCTCGCGACCCTGACCATGGTCTGGCCGACAGGTACACGTCGCGATTTTTCTTGAAGCATTCTTTCATCGAGCAGGCTTTCGACATGTTGCAAGAGAACGGATTCAAACTGACGGCCAGCTGTGGCAGCGGGACCGCCGGTGGAGCCGCCGAACTGAAACCCGGCGTCGACTCTGAAGAGAATCGTTGGAACCATTACAACGAATTTGTGTTCGTGCGTGAATAG
Protein
MGSRGAETCGKWVREGAGRAAAATLRPQPLTMADAPPEVLELNVGGVHYATTRDTLLREPDSLPAAALQDPDHPRDARGRIFFDRDGVLFRYVLDYLRDGGLVLPECFREHKRLAREAKHYRLVGLENAVREADPRPPNRERGCITVGYRGSFAFGRDGLADVKFRKLSRILVCGKVTLCRDVFGETLNESRDPDHGLADRYTSRFFLKHSFIEQAFDMLQENGFKLTASCGSGTAGGAAELKPGVDSEENRWNHYNEFVFVRE
Summary
Uniprot
A0A212FLH7
A0A2A4IXI0
A0A194PL85
H9JPH0
A0A0L7K406
A0A1Y1LHD9
+ More
A0A0C9R4Q7 A0A1J1IXS1 A0A067RHN4 A0A2J7RD91 A0A1W4WTS7 A0A2P8XHK2 A0A1B0DK36 A0A1B0FK32 A0A1A9X5X5 A0A1A9Z7D2 D6W8K7 A0A1A9V4Y8 A0A1L8DCM9 A0A1B0AWL9 E2BH63 A0A1A9W568 E0VD68 A0A0A1XDN3 E9IIW0 A0A2A3ET42 V9IJR9 A0A088ACU7 A0A195D3I7 A0A0J7NKZ2 A0A0L0BY39 F4WWQ4 A0A151JNN5 E1ZWV3 A0A195BLG4 A0A158NJ09 A0A310SN01 A0A1B6LBP6 K7IR02 A0A0L7QRA3 A0A195FHG1 A0A026WKE1 A0A1I8PX01 B4KBF1 U5EF17 W8BGF3 B4MB74 A0A1S3DBQ2 A0A1I8M3Q1 A0A1B6J142 A0A1B6FSB2 A0A1W4VAF0 B3M355 B4R1Q0 B4II53 A0A0M4EDW2 Q9VDH3 B4NFR6 A0A3B0K0B1 Q299E5 B4G531 B4PQ89 A0A336KR81 B3P310 T1HPM3 U4ULH4 A0A146KJJ7 A0A1B0CW58 A0A0A9W9B5 A0A2S2QKI6 A0A2H8TF44 A0A2S2P2T6 J9JYM6 B4JSC6 A0A182R0K3 A0A182VL94 Q7QE64 E9H8K1 A0A0P6H8X1 A0A162S4I1 A0A182I9N7 A0A182TSV6 A0A182X599 A0A182P833 A0A0L8G5C0 W5JMF5 A0A182NVQ0 A0A023F559 A0A182FK08 A0A182JAY6 A0A182KHH9 A0A084WPJ7 A0A182MNP4 T1IPD2 A0A2R7VY57 Q16YX0 A0A182GGU7 A0A182VV97 R7VBA0 A0A182G9Q6
A0A0C9R4Q7 A0A1J1IXS1 A0A067RHN4 A0A2J7RD91 A0A1W4WTS7 A0A2P8XHK2 A0A1B0DK36 A0A1B0FK32 A0A1A9X5X5 A0A1A9Z7D2 D6W8K7 A0A1A9V4Y8 A0A1L8DCM9 A0A1B0AWL9 E2BH63 A0A1A9W568 E0VD68 A0A0A1XDN3 E9IIW0 A0A2A3ET42 V9IJR9 A0A088ACU7 A0A195D3I7 A0A0J7NKZ2 A0A0L0BY39 F4WWQ4 A0A151JNN5 E1ZWV3 A0A195BLG4 A0A158NJ09 A0A310SN01 A0A1B6LBP6 K7IR02 A0A0L7QRA3 A0A195FHG1 A0A026WKE1 A0A1I8PX01 B4KBF1 U5EF17 W8BGF3 B4MB74 A0A1S3DBQ2 A0A1I8M3Q1 A0A1B6J142 A0A1B6FSB2 A0A1W4VAF0 B3M355 B4R1Q0 B4II53 A0A0M4EDW2 Q9VDH3 B4NFR6 A0A3B0K0B1 Q299E5 B4G531 B4PQ89 A0A336KR81 B3P310 T1HPM3 U4ULH4 A0A146KJJ7 A0A1B0CW58 A0A0A9W9B5 A0A2S2QKI6 A0A2H8TF44 A0A2S2P2T6 J9JYM6 B4JSC6 A0A182R0K3 A0A182VL94 Q7QE64 E9H8K1 A0A0P6H8X1 A0A162S4I1 A0A182I9N7 A0A182TSV6 A0A182X599 A0A182P833 A0A0L8G5C0 W5JMF5 A0A182NVQ0 A0A023F559 A0A182FK08 A0A182JAY6 A0A182KHH9 A0A084WPJ7 A0A182MNP4 T1IPD2 A0A2R7VY57 Q16YX0 A0A182GGU7 A0A182VV97 R7VBA0 A0A182G9Q6
Pubmed
22118469
26354079
19121390
26227816
28004739
24845553
+ More
29403074 18362917 19820115 20798317 20566863 25830018 21282665 26108605 21719571 21347285 20075255 24508170 30249741 17994087 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 23537049 26823975 25401762 12364791 14747013 17210077 21292972 20920257 23761445 25474469 24438588 17510324 26483478 23254933
29403074 18362917 19820115 20798317 20566863 25830018 21282665 26108605 21719571 21347285 20075255 24508170 30249741 17994087 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 23537049 26823975 25401762 12364791 14747013 17210077 21292972 20920257 23761445 25474469 24438588 17510324 26483478 23254933
EMBL
AGBW02007799
OWR54549.1
NWSH01005123
PCG64421.1
KQ459601
KPI94087.1
+ More
BABH01015560 JTDY01011169 KOB57437.1 GEZM01059725 JAV71385.1 GBYB01002090 GBYB01002950 GBYB01002951 JAG71857.1 JAG72717.1 JAG72718.1 CVRI01000063 CRL04880.1 KK852463 KDR23376.1 NEVH01005293 PNF38788.1 PYGN01002102 PSN31488.1 AJVK01015896 CCAG010014364 KQ971312 EEZ98371.1 GFDF01009865 JAV04219.1 JXJN01004816 GL448268 EFN84984.1 DS235072 EEB11324.1 GBXI01005220 JAD09072.1 GL763562 EFZ19482.1 KZ288185 PBC34953.1 JR048671 AEY60726.1 KQ976885 KYN07462.1 LBMM01003811 KMQ93110.1 JRES01001168 KNC24886.1 GL888412 EGI61376.1 KQ978889 KYN27663.1 GL434905 EFN74350.1 KQ976453 KYM85517.1 ADTU01017348 KQ759841 OAD62625.1 GEBQ01018868 JAT21109.1 KQ414782 KOC61157.1 KQ981523 KYN40135.1 KK107163 QOIP01000002 EZA56522.1 RLU25513.1 CH933806 EDW16879.1 GANO01004062 JAB55809.1 GAMC01014195 GAMC01014194 GAMC01014193 JAB92361.1 CH940656 EDW58345.1 GECU01014876 JAS92830.1 GECZ01020857 GECZ01016715 GECZ01011657 GECZ01005140 JAS48912.1 JAS53054.1 JAS58112.1 JAS64629.1 CH902617 EDV42455.1 CM000364 EDX12159.1 CH480842 EDW49597.1 CP012526 ALC46078.1 AE014297 AY060259 AAF55820.1 AAL25298.1 ALI30594.1 CH964251 EDW83133.1 OUUW01000013 SPP87747.1 CM000070 EAL27758.1 CH479179 EDW24697.1 CM000160 EDW96198.1 UFQS01000433 UFQS01000851 UFQT01000433 UFQT01000851 SSX03895.1 SSX07304.1 SSX24260.1 CH954181 EDV48324.1 ACPB03012029 KB632401 ERL94919.1 GDHC01021868 JAP96760.1 AJWK01031788 AJWK01031789 AJWK01031790 GBHO01038572 JAG05032.1 GGMS01009025 MBY78228.1 GFXV01000103 MBW11908.1 GGMR01011158 MBY23777.1 ABLF02039240 CH916373 EDV94666.1 AXCN02001376 AAAB01008847 EAA06845.3 GL732605 EFX71930.1 GDIQ01023041 JAN71696.1 LRGB01000084 KZS21007.1 APCN01001014 KQ423864 KOF72049.1 ADMH02000996 ETN64483.1 GBBI01002504 JAC16208.1 AXCP01009642 ATLV01025083 KE525369 KFB52141.1 AXCM01016581 JH431256 KK854165 PTY12434.1 CH477507 EAT39818.1 JXUM01062431 KQ562199 KXJ76428.1 AMQN01019304 KB295573 ELU12980.1 JXUM01008283 KQ560257 KXJ83511.1
BABH01015560 JTDY01011169 KOB57437.1 GEZM01059725 JAV71385.1 GBYB01002090 GBYB01002950 GBYB01002951 JAG71857.1 JAG72717.1 JAG72718.1 CVRI01000063 CRL04880.1 KK852463 KDR23376.1 NEVH01005293 PNF38788.1 PYGN01002102 PSN31488.1 AJVK01015896 CCAG010014364 KQ971312 EEZ98371.1 GFDF01009865 JAV04219.1 JXJN01004816 GL448268 EFN84984.1 DS235072 EEB11324.1 GBXI01005220 JAD09072.1 GL763562 EFZ19482.1 KZ288185 PBC34953.1 JR048671 AEY60726.1 KQ976885 KYN07462.1 LBMM01003811 KMQ93110.1 JRES01001168 KNC24886.1 GL888412 EGI61376.1 KQ978889 KYN27663.1 GL434905 EFN74350.1 KQ976453 KYM85517.1 ADTU01017348 KQ759841 OAD62625.1 GEBQ01018868 JAT21109.1 KQ414782 KOC61157.1 KQ981523 KYN40135.1 KK107163 QOIP01000002 EZA56522.1 RLU25513.1 CH933806 EDW16879.1 GANO01004062 JAB55809.1 GAMC01014195 GAMC01014194 GAMC01014193 JAB92361.1 CH940656 EDW58345.1 GECU01014876 JAS92830.1 GECZ01020857 GECZ01016715 GECZ01011657 GECZ01005140 JAS48912.1 JAS53054.1 JAS58112.1 JAS64629.1 CH902617 EDV42455.1 CM000364 EDX12159.1 CH480842 EDW49597.1 CP012526 ALC46078.1 AE014297 AY060259 AAF55820.1 AAL25298.1 ALI30594.1 CH964251 EDW83133.1 OUUW01000013 SPP87747.1 CM000070 EAL27758.1 CH479179 EDW24697.1 CM000160 EDW96198.1 UFQS01000433 UFQS01000851 UFQT01000433 UFQT01000851 SSX03895.1 SSX07304.1 SSX24260.1 CH954181 EDV48324.1 ACPB03012029 KB632401 ERL94919.1 GDHC01021868 JAP96760.1 AJWK01031788 AJWK01031789 AJWK01031790 GBHO01038572 JAG05032.1 GGMS01009025 MBY78228.1 GFXV01000103 MBW11908.1 GGMR01011158 MBY23777.1 ABLF02039240 CH916373 EDV94666.1 AXCN02001376 AAAB01008847 EAA06845.3 GL732605 EFX71930.1 GDIQ01023041 JAN71696.1 LRGB01000084 KZS21007.1 APCN01001014 KQ423864 KOF72049.1 ADMH02000996 ETN64483.1 GBBI01002504 JAC16208.1 AXCP01009642 ATLV01025083 KE525369 KFB52141.1 AXCM01016581 JH431256 KK854165 PTY12434.1 CH477507 EAT39818.1 JXUM01062431 KQ562199 KXJ76428.1 AMQN01019304 KB295573 ELU12980.1 JXUM01008283 KQ560257 KXJ83511.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000005204
UP000037510
UP000183832
+ More
UP000027135 UP000235965 UP000192223 UP000245037 UP000092462 UP000092444 UP000092443 UP000092445 UP000007266 UP000078200 UP000092460 UP000008237 UP000091820 UP000009046 UP000242457 UP000005203 UP000078542 UP000036403 UP000037069 UP000007755 UP000078492 UP000000311 UP000078540 UP000005205 UP000002358 UP000053825 UP000078541 UP000053097 UP000279307 UP000095300 UP000009192 UP000008792 UP000079169 UP000095301 UP000192221 UP000007801 UP000000304 UP000001292 UP000092553 UP000000803 UP000007798 UP000268350 UP000001819 UP000008744 UP000002282 UP000008711 UP000015103 UP000030742 UP000092461 UP000007819 UP000001070 UP000075886 UP000075903 UP000007062 UP000000305 UP000076858 UP000075840 UP000075902 UP000076407 UP000075885 UP000053454 UP000000673 UP000075884 UP000069272 UP000075880 UP000075881 UP000030765 UP000075883 UP000008820 UP000069940 UP000249989 UP000075920 UP000014760
UP000027135 UP000235965 UP000192223 UP000245037 UP000092462 UP000092444 UP000092443 UP000092445 UP000007266 UP000078200 UP000092460 UP000008237 UP000091820 UP000009046 UP000242457 UP000005203 UP000078542 UP000036403 UP000037069 UP000007755 UP000078492 UP000000311 UP000078540 UP000005205 UP000002358 UP000053825 UP000078541 UP000053097 UP000279307 UP000095300 UP000009192 UP000008792 UP000079169 UP000095301 UP000192221 UP000007801 UP000000304 UP000001292 UP000092553 UP000000803 UP000007798 UP000268350 UP000001819 UP000008744 UP000002282 UP000008711 UP000015103 UP000030742 UP000092461 UP000007819 UP000001070 UP000075886 UP000075903 UP000007062 UP000000305 UP000076858 UP000075840 UP000075902 UP000076407 UP000075885 UP000053454 UP000000673 UP000075884 UP000069272 UP000075880 UP000075881 UP000030765 UP000075883 UP000008820 UP000069940 UP000249989 UP000075920 UP000014760
Pfam
PF02214 BTB_2
SUPFAM
SSF54695
SSF54695
ProteinModelPortal
A0A212FLH7
A0A2A4IXI0
A0A194PL85
H9JPH0
A0A0L7K406
A0A1Y1LHD9
+ More
A0A0C9R4Q7 A0A1J1IXS1 A0A067RHN4 A0A2J7RD91 A0A1W4WTS7 A0A2P8XHK2 A0A1B0DK36 A0A1B0FK32 A0A1A9X5X5 A0A1A9Z7D2 D6W8K7 A0A1A9V4Y8 A0A1L8DCM9 A0A1B0AWL9 E2BH63 A0A1A9W568 E0VD68 A0A0A1XDN3 E9IIW0 A0A2A3ET42 V9IJR9 A0A088ACU7 A0A195D3I7 A0A0J7NKZ2 A0A0L0BY39 F4WWQ4 A0A151JNN5 E1ZWV3 A0A195BLG4 A0A158NJ09 A0A310SN01 A0A1B6LBP6 K7IR02 A0A0L7QRA3 A0A195FHG1 A0A026WKE1 A0A1I8PX01 B4KBF1 U5EF17 W8BGF3 B4MB74 A0A1S3DBQ2 A0A1I8M3Q1 A0A1B6J142 A0A1B6FSB2 A0A1W4VAF0 B3M355 B4R1Q0 B4II53 A0A0M4EDW2 Q9VDH3 B4NFR6 A0A3B0K0B1 Q299E5 B4G531 B4PQ89 A0A336KR81 B3P310 T1HPM3 U4ULH4 A0A146KJJ7 A0A1B0CW58 A0A0A9W9B5 A0A2S2QKI6 A0A2H8TF44 A0A2S2P2T6 J9JYM6 B4JSC6 A0A182R0K3 A0A182VL94 Q7QE64 E9H8K1 A0A0P6H8X1 A0A162S4I1 A0A182I9N7 A0A182TSV6 A0A182X599 A0A182P833 A0A0L8G5C0 W5JMF5 A0A182NVQ0 A0A023F559 A0A182FK08 A0A182JAY6 A0A182KHH9 A0A084WPJ7 A0A182MNP4 T1IPD2 A0A2R7VY57 Q16YX0 A0A182GGU7 A0A182VV97 R7VBA0 A0A182G9Q6
A0A0C9R4Q7 A0A1J1IXS1 A0A067RHN4 A0A2J7RD91 A0A1W4WTS7 A0A2P8XHK2 A0A1B0DK36 A0A1B0FK32 A0A1A9X5X5 A0A1A9Z7D2 D6W8K7 A0A1A9V4Y8 A0A1L8DCM9 A0A1B0AWL9 E2BH63 A0A1A9W568 E0VD68 A0A0A1XDN3 E9IIW0 A0A2A3ET42 V9IJR9 A0A088ACU7 A0A195D3I7 A0A0J7NKZ2 A0A0L0BY39 F4WWQ4 A0A151JNN5 E1ZWV3 A0A195BLG4 A0A158NJ09 A0A310SN01 A0A1B6LBP6 K7IR02 A0A0L7QRA3 A0A195FHG1 A0A026WKE1 A0A1I8PX01 B4KBF1 U5EF17 W8BGF3 B4MB74 A0A1S3DBQ2 A0A1I8M3Q1 A0A1B6J142 A0A1B6FSB2 A0A1W4VAF0 B3M355 B4R1Q0 B4II53 A0A0M4EDW2 Q9VDH3 B4NFR6 A0A3B0K0B1 Q299E5 B4G531 B4PQ89 A0A336KR81 B3P310 T1HPM3 U4ULH4 A0A146KJJ7 A0A1B0CW58 A0A0A9W9B5 A0A2S2QKI6 A0A2H8TF44 A0A2S2P2T6 J9JYM6 B4JSC6 A0A182R0K3 A0A182VL94 Q7QE64 E9H8K1 A0A0P6H8X1 A0A162S4I1 A0A182I9N7 A0A182TSV6 A0A182X599 A0A182P833 A0A0L8G5C0 W5JMF5 A0A182NVQ0 A0A023F559 A0A182FK08 A0A182JAY6 A0A182KHH9 A0A084WPJ7 A0A182MNP4 T1IPD2 A0A2R7VY57 Q16YX0 A0A182GGU7 A0A182VV97 R7VBA0 A0A182G9Q6
PDB
6QB7
E-value=1.51629e-28,
Score=312
Ontologies
GO
Topology
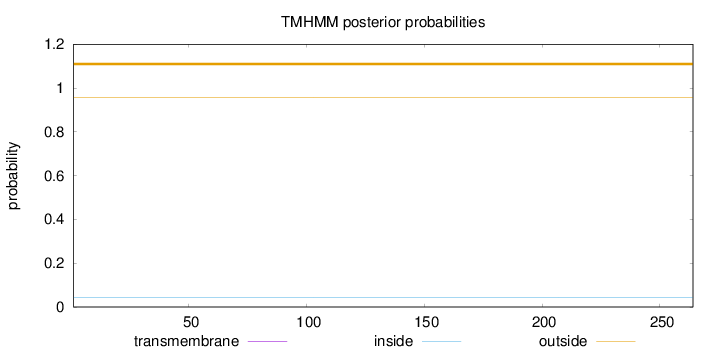
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000710000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04289
outside
1 - 264
Population Genetic Test Statistics
Pi
156.456758
Theta
156.713198
Tajima's D
0.018558
CLR
0.608825
CSRT
0.378381080945953
Interpretation
Uncertain
