Pre Gene Modal
BGIBMGA011426
Annotation
PREDICTED:_LETM1_and_EF-hand_domain-containing_protein_1?_mitochondrial_[Bombyx_mori]
Full name
Mitochondrial proton/calcium exchanger protein
Alternative Name
Leucine zipper-EF-hand-containing transmembrane protein 1
Location in the cell
Cytoplasmic Reliability : 1.421 Mitochondrial Reliability : 1.693 Nuclear Reliability : 1.174
Sequence
CDS
ATGAACAGAATAATTTTAAATCGATGCCGGTTTATAAAAAAATCAACAATATTATGGGAGACAAGACAGTATCAAAATTTTCCATACATAACATTGTCAAGGTCACTATCGTACTACACAGCACAGCCGATAGGCCTCCAACACGATGTCACACAGAAACCTCTCAGCATGCAGGTGTACTATCCTACATTTACCAGACAATTTTCCACAAGTCAGTATTATTTTGAGAAAGAACCATTAAAACCTTCCTCAAAAGTAGAAGCTACTGTTGTTACAATTAAAAAAGAAATAGAGGAGAAAGAAAAACTCCCTAAAAAAGAAGAGAGAAAGAAGAGTATCAAGGAACGTGTTATGGATGAACTTCGCCATTATTACCATGGCTTCAGGTTGCTTTTCATAGAAACTAGAATATCAATTTCATTGTTATTTAGAATATTAAAAGGAGATACATTAACTAGGAGGGAACATAGACTGCTAGTGACAACAGTTGGTGATCTGTTTAGAATGGTGCCCTTCATCATATTTATAATAGTTCCATTCTTAGAATTCTTTTTACCAGTATTCATAAAGCTTTTTCCCAATATGTTACCATCAACATTTGAAAGTAGTAGTCAAAAAGAAGCAAAACTGAAGCAACATTTGAAAGTCAAACTAGAAATGGCCAAGTTTTTTCAAGAAACATTAGATCAGATGGCACCACAAGCTAGTAATCGTCATTCTGAATTAGCTAAAGAGTTTTCAAATTTCTTTAGTAGAATAAGAACATCAGGGGATGTGGCAACAAGTGAAGAGATTATGAAATTTTCAAAGTTGTTTGAAGATGAAATCACCCTTGATTCTCTCCAGAGACCACACCTTGTTGCTCTTTGCAAAGTTTTAAATGTTTCTACAATTGGTACTAGTGCAATGTTAAGGTTTAATCTCAAAATGAAGTTAAGGTCACTAGCGGCAGATGACAAAATGATAGCTAAAGAAGGTGTGGATAGTTTGAACTTTAGTGAACTACAACAAGCTTGTAGAGCACGAGGAATGAGAGCATATGGTGTGTCTGAAGAGCGCTTACGTAAAGAACTGAGGAATTGGTTAGATTTATCACTGAATGAAAAGGTTCCCCCTTCGTTGTTGTTGTTGTCAAGAGCTCTGATGGTACCGGAGCATGTACCAACAACATATAAGCTGAAGGCTACAATCTCAGCTTTGCCAGAACATGTTGCAACACAAACTAAAGCTGCAATCGGAGAAAAGGAAGGCAAACTTGATTTTAAGGCCAAAGCAGAAGCAATCAGACTTGAAGAAATGAAGATTAAGGAAGAGAAACAAGAACTCCAAGAGGCAGAAAGGGAGAAGCAACTGCTTGAAGCCAAGAAGAGAGAGAAGGAGGAATTGAAAGACAAAGCACCCATCATTGATGCTGAGTCAACTCCAATTTTAGTTGATCCAGCTCCAGTGATTGCAGTTGACCCACCCAAACAGAAGTCTGAAGAAGGTTTATCTGGCAAGGACATTGAAGTTATTGAAGACGCTTTGGAAAAATTGGCTGAGCAGAAGAAGTCCTTGCTTCTCGAGAAGGAAAGCATTCAGGAGCTAAAAACGGAACTGTTGGACTACAGCGAAGATGTGAAAGAGATGCACGAGATTGTCAAGACAAAGCAAGCCGATATCAAACTTACGAAAGGTGCACGAAGATTATATCGAGCTGTCAATAACATGATAAGCAAACTTGACGCTGCCCTGGTGGAGCTTGAAAGCAAAGAGAAAGCCTTGAAGACGACACTAGAACAGACCAAGTCCGAACCACAGAAAGCTAAAGAACAGTTGATCCAAATCGACGAACTGATGCATTCCATTAAGAGCTTACAAAAAGTCCCCGATGAGGCTAAGTTGAAGTTAATTCAAGACGTACTGGGCAGGCTAGACGACGACTTTGATGGACAGTTGAAGATTGATGATGTTTTGAAGATGCTCGAAATAATCGGTAACGAAAACGTGAATCTATCAGAGAAACAGATGGCCGAATTAATCGAACTACTCGACAAAGAGGAGATTCTCGAGGTCGAAAGCAAGATACAGAAGGCGTTAGATAAAGCCGCCAGTGCCGCCAAAGAACAGGACAAGAACAGTGAATCCGATGGAAAGAGTCTATTCGATATCATAAGGGAGGCCCAGAAGCGAAGAGAAATGAAAAAAGCAGTCGAGAAATTCGGAGAACAAGGAGATATCCCTAAGAGTGACGTCATCGCGAAAACCGAGGAGAGCAGTAAACGGCCGGAGAGCCACTGA
Protein
MNRIILNRCRFIKKSTILWETRQYQNFPYITLSRSLSYYTAQPIGLQHDVTQKPLSMQVYYPTFTRQFSTSQYYFEKEPLKPSSKVEATVVTIKKEIEEKEKLPKKEERKKSIKERVMDELRHYYHGFRLLFIETRISISLLFRILKGDTLTRREHRLLVTTVGDLFRMVPFIIFIIVPFLEFFLPVFIKLFPNMLPSTFESSSQKEAKLKQHLKVKLEMAKFFQETLDQMAPQASNRHSELAKEFSNFFSRIRTSGDVATSEEIMKFSKLFEDEITLDSLQRPHLVALCKVLNVSTIGTSAMLRFNLKMKLRSLAADDKMIAKEGVDSLNFSELQQACRARGMRAYGVSEERLRKELRNWLDLSLNEKVPPSLLLLSRALMVPEHVPTTYKLKATISALPEHVATQTKAAIGEKEGKLDFKAKAEAIRLEEMKIKEEKQELQEAEREKQLLEAKKREKEELKDKAPIIDAESTPILVDPAPVIAVDPPKQKSEEGLSGKDIEVIEDALEKLAEQKKSLLLEKESIQELKTELLDYSEDVKEMHEIVKTKQADIKLTKGARRLYRAVNNMISKLDAALVELESKEKALKTTLEQTKSEPQKAKEQLIQIDELMHSIKSLQKVPDEAKLKLIQDVLGRLDDDFDGQLKIDDVLKMLEIIGNENVNLSEKQMAELIELLDKEEILEVESKIQKALDKAASAAKEQDKNSESDGKSLFDIIREAQKRREMKKAVEKFGEQGDIPKSDVIAKTEESSKRPESH
Summary
Description
Mitochondrial proton/calcium antiporter that mediates proton-dependent calcium efflux from mitochondrion (By similarity). Required for the maintenance of the tubular shape and cristae organization (By similarity).
Mitochondrial proton/calcium antiporter that mediates proton-dependent calcium efflux from mitochondrion (By similarity). Crucial for the maintenance of mitochondrial tubular networks and for the assembly of the supercomplexes of the respiratory chain (By similarity). Required for the maintenance of the tubular shape and cristae organization (By similarity). In contrast to SLC8B1/NCLX, does not constitute the major factor for mitochondrial calcium extrusion (By similarity).
Mitochondrial proton/calcium antiporter that mediates proton-dependent calcium efflux from mitochondrion (By similarity). Crucial for the maintenance of mitochondrial tubular networks and for the assembly of the supercomplexes of the respiratory chain (By similarity). Required for the maintenance of the tubular shape and cristae organization (By similarity). In contrast to SLC8B1/NCLX, does not constitute the major factor for mitochondrial calcium extrusion (By similarity).
Subunit
Homohexamer.
Homohexamer (By similarity). Interacts with BCS1L (By similarity).
Homohexamer (By similarity). Interacts with BCS1L (By similarity).
Similarity
Belongs to the LETM1 family.
Keywords
Antiport
Calcium
Calcium transport
Coiled coil
Complete proteome
Ion transport
Membrane
Metal-binding
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transmembrane
Transmembrane helix
Transport
Feature
chain Mitochondrial proton/calcium exchanger protein
Uniprot
A0A194PSI7
A0A212FLE1
A0A3S2NZL5
A0A2H1VBJ8
A0A1E1WBU9
A0A2A4JF37
+ More
A0A0L7L5Q4 A0A194RH62 H9JPH1 D6WBJ8 A0A0T6B418 A0A1L8E357 A0A1L8E358 V5GTP2 A0A1W4X5U9 A0A1B6M0Q3 A0A1B6M0V2 U4U5U4 N6T685 A0A195F3V7 A0A146KQC4 A0A2J7QSB1 F4WZ28 R4WTU9 E0VJ68 U5EUZ5 A0A026W0U3 A0A067R7R2 E2C5E1 A0A2H8TK42 A0A0L7RBT9 A0A2H8U2I8 A0A0C9R2H2 J9JYY9 A0A2S2PV58 A0A2S2R4N6 A0A0K8V865 A0A0P4YN60 A0A0N8B814 A0A087TWM2 A0A0P5J4L6 A0A0P5KX56 A0A0A1WWG5 A0A1B6HUG7 A0A0P5FSM7 A0A0N8CYY7 A0A1Y3AQ25 E9FQN6 A0A0P6CME8 W8BGG6 L7M528 A0A1A9Z7M9 A0A224YSK6 A0A1A9WBX2 A0A131YQF5 A0A1B0AP14 A0A1A9XVX4 A0A1A9VIC2 B4LKF7 A0A131XMB7 A0A091PWY7 A0A091QLX0 A0A1I7Q423 Q5ZK33 A0A091K1H7 E9GYB1 A0A226MWV8 A0A0P5LN53 A0A3Q1MMS8 G3U258 L8IIK8 Q0VCA3 F1N3R4 A0A2K6LZ68 A0A2K6QN82 K9INF8
A0A0L7L5Q4 A0A194RH62 H9JPH1 D6WBJ8 A0A0T6B418 A0A1L8E357 A0A1L8E358 V5GTP2 A0A1W4X5U9 A0A1B6M0Q3 A0A1B6M0V2 U4U5U4 N6T685 A0A195F3V7 A0A146KQC4 A0A2J7QSB1 F4WZ28 R4WTU9 E0VJ68 U5EUZ5 A0A026W0U3 A0A067R7R2 E2C5E1 A0A2H8TK42 A0A0L7RBT9 A0A2H8U2I8 A0A0C9R2H2 J9JYY9 A0A2S2PV58 A0A2S2R4N6 A0A0K8V865 A0A0P4YN60 A0A0N8B814 A0A087TWM2 A0A0P5J4L6 A0A0P5KX56 A0A0A1WWG5 A0A1B6HUG7 A0A0P5FSM7 A0A0N8CYY7 A0A1Y3AQ25 E9FQN6 A0A0P6CME8 W8BGG6 L7M528 A0A1A9Z7M9 A0A224YSK6 A0A1A9WBX2 A0A131YQF5 A0A1B0AP14 A0A1A9XVX4 A0A1A9VIC2 B4LKF7 A0A131XMB7 A0A091PWY7 A0A091QLX0 A0A1I7Q423 Q5ZK33 A0A091K1H7 E9GYB1 A0A226MWV8 A0A0P5LN53 A0A3Q1MMS8 G3U258 L8IIK8 Q0VCA3 F1N3R4 A0A2K6LZ68 A0A2K6QN82 K9INF8
Pubmed
EMBL
KQ459601
KPI94085.1
AGBW02007799
OWR54547.1
RSAL01000081
RVE48577.1
+ More
ODYU01001652 SOQ38176.1 GDQN01006703 JAT84351.1 NWSH01001810 PCG70053.1 JTDY01002720 KOB70853.1 KQ460205 KPJ17163.1 BABH01015558 KQ971311 EEZ98994.2 LJIG01009909 KRT82138.1 GFDF01000926 JAV13158.1 GFDF01000927 JAV13157.1 GALX01003469 JAB64997.1 GEBQ01010517 JAT29460.1 GEBQ01010463 JAT29514.1 KB631984 ERL87713.1 APGK01042111 KB741002 ENN75724.1 KQ981820 KYN35143.1 GDHC01020744 JAP97884.1 NEVH01011876 PNF31479.1 GL888465 EGI60531.1 AK418132 BAN21347.1 DS235219 EEB13424.1 GANO01003643 JAB56228.1 KK107578 QOIP01000002 EZA48684.1 RLU25684.1 KK852643 KDR19565.1 GL452770 EFN76874.1 GFXV01002595 MBW14400.1 KQ414617 KOC68270.1 GFXV01007943 MBW19748.1 GBYB01010279 JAG80046.1 ABLF02015848 GGMS01000106 MBY69309.1 GGMS01015752 MBY84955.1 GDHF01017182 GDHF01014886 GDHF01010754 GDHF01009064 GDHF01005594 GDHF01001993 JAI35132.1 JAI37428.1 JAI41560.1 JAI43250.1 JAI46720.1 JAI50321.1 GDIP01225068 JAI98333.1 GDIQ01203747 JAK47978.1 KK117088 KFM69511.1 GDIQ01205399 JAK46326.1 GDIQ01203746 JAK47979.1 GBXI01010898 JAD03394.1 GECU01029394 JAS78312.1 GDIP01144270 JAJ79132.1 GDIP01085151 JAM18564.1 MUJZ01065227 OTF70549.1 GL732523 EFX90017.1 GDIP01012470 JAM91245.1 GAMC01010528 GAMC01010527 GAMC01010526 JAB96028.1 GACK01006840 JAA58194.1 GFPF01009420 MAA20566.1 GEDV01007390 JAP81167.1 JXJN01001110 CH940648 EDW60678.1 KRF79537.1 KRF79538.1 GEFH01001910 JAP66671.1 KK680568 KFQ12452.1 KK799412 KFQ27621.1 AADN05000017 AJ720251 KK538502 KFP30141.1 GL732574 EFX75571.1 MCFN01000373 OXB59761.1 GDIQ01167452 JAK84273.1 JH881175 ELR55983.1 BC120274 GABZ01004638 JAA48887.1
ODYU01001652 SOQ38176.1 GDQN01006703 JAT84351.1 NWSH01001810 PCG70053.1 JTDY01002720 KOB70853.1 KQ460205 KPJ17163.1 BABH01015558 KQ971311 EEZ98994.2 LJIG01009909 KRT82138.1 GFDF01000926 JAV13158.1 GFDF01000927 JAV13157.1 GALX01003469 JAB64997.1 GEBQ01010517 JAT29460.1 GEBQ01010463 JAT29514.1 KB631984 ERL87713.1 APGK01042111 KB741002 ENN75724.1 KQ981820 KYN35143.1 GDHC01020744 JAP97884.1 NEVH01011876 PNF31479.1 GL888465 EGI60531.1 AK418132 BAN21347.1 DS235219 EEB13424.1 GANO01003643 JAB56228.1 KK107578 QOIP01000002 EZA48684.1 RLU25684.1 KK852643 KDR19565.1 GL452770 EFN76874.1 GFXV01002595 MBW14400.1 KQ414617 KOC68270.1 GFXV01007943 MBW19748.1 GBYB01010279 JAG80046.1 ABLF02015848 GGMS01000106 MBY69309.1 GGMS01015752 MBY84955.1 GDHF01017182 GDHF01014886 GDHF01010754 GDHF01009064 GDHF01005594 GDHF01001993 JAI35132.1 JAI37428.1 JAI41560.1 JAI43250.1 JAI46720.1 JAI50321.1 GDIP01225068 JAI98333.1 GDIQ01203747 JAK47978.1 KK117088 KFM69511.1 GDIQ01205399 JAK46326.1 GDIQ01203746 JAK47979.1 GBXI01010898 JAD03394.1 GECU01029394 JAS78312.1 GDIP01144270 JAJ79132.1 GDIP01085151 JAM18564.1 MUJZ01065227 OTF70549.1 GL732523 EFX90017.1 GDIP01012470 JAM91245.1 GAMC01010528 GAMC01010527 GAMC01010526 JAB96028.1 GACK01006840 JAA58194.1 GFPF01009420 MAA20566.1 GEDV01007390 JAP81167.1 JXJN01001110 CH940648 EDW60678.1 KRF79537.1 KRF79538.1 GEFH01001910 JAP66671.1 KK680568 KFQ12452.1 KK799412 KFQ27621.1 AADN05000017 AJ720251 KK538502 KFP30141.1 GL732574 EFX75571.1 MCFN01000373 OXB59761.1 GDIQ01167452 JAK84273.1 JH881175 ELR55983.1 BC120274 GABZ01004638 JAA48887.1
Proteomes
UP000053268
UP000007151
UP000283053
UP000218220
UP000037510
UP000053240
+ More
UP000005204 UP000007266 UP000192223 UP000030742 UP000019118 UP000078541 UP000235965 UP000007755 UP000009046 UP000053097 UP000279307 UP000027135 UP000008237 UP000053825 UP000007819 UP000054359 UP000000305 UP000092445 UP000091820 UP000092460 UP000092443 UP000078200 UP000008792 UP000000539 UP000198323 UP000009136 UP000007646 UP000233180 UP000233200
UP000005204 UP000007266 UP000192223 UP000030742 UP000019118 UP000078541 UP000235965 UP000007755 UP000009046 UP000053097 UP000279307 UP000027135 UP000008237 UP000053825 UP000007819 UP000054359 UP000000305 UP000092445 UP000091820 UP000092460 UP000092443 UP000078200 UP000008792 UP000000539 UP000198323 UP000009136 UP000007646 UP000233180 UP000233200
Interpro
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
A0A194PSI7
A0A212FLE1
A0A3S2NZL5
A0A2H1VBJ8
A0A1E1WBU9
A0A2A4JF37
+ More
A0A0L7L5Q4 A0A194RH62 H9JPH1 D6WBJ8 A0A0T6B418 A0A1L8E357 A0A1L8E358 V5GTP2 A0A1W4X5U9 A0A1B6M0Q3 A0A1B6M0V2 U4U5U4 N6T685 A0A195F3V7 A0A146KQC4 A0A2J7QSB1 F4WZ28 R4WTU9 E0VJ68 U5EUZ5 A0A026W0U3 A0A067R7R2 E2C5E1 A0A2H8TK42 A0A0L7RBT9 A0A2H8U2I8 A0A0C9R2H2 J9JYY9 A0A2S2PV58 A0A2S2R4N6 A0A0K8V865 A0A0P4YN60 A0A0N8B814 A0A087TWM2 A0A0P5J4L6 A0A0P5KX56 A0A0A1WWG5 A0A1B6HUG7 A0A0P5FSM7 A0A0N8CYY7 A0A1Y3AQ25 E9FQN6 A0A0P6CME8 W8BGG6 L7M528 A0A1A9Z7M9 A0A224YSK6 A0A1A9WBX2 A0A131YQF5 A0A1B0AP14 A0A1A9XVX4 A0A1A9VIC2 B4LKF7 A0A131XMB7 A0A091PWY7 A0A091QLX0 A0A1I7Q423 Q5ZK33 A0A091K1H7 E9GYB1 A0A226MWV8 A0A0P5LN53 A0A3Q1MMS8 G3U258 L8IIK8 Q0VCA3 F1N3R4 A0A2K6LZ68 A0A2K6QN82 K9INF8
A0A0L7L5Q4 A0A194RH62 H9JPH1 D6WBJ8 A0A0T6B418 A0A1L8E357 A0A1L8E358 V5GTP2 A0A1W4X5U9 A0A1B6M0Q3 A0A1B6M0V2 U4U5U4 N6T685 A0A195F3V7 A0A146KQC4 A0A2J7QSB1 F4WZ28 R4WTU9 E0VJ68 U5EUZ5 A0A026W0U3 A0A067R7R2 E2C5E1 A0A2H8TK42 A0A0L7RBT9 A0A2H8U2I8 A0A0C9R2H2 J9JYY9 A0A2S2PV58 A0A2S2R4N6 A0A0K8V865 A0A0P4YN60 A0A0N8B814 A0A087TWM2 A0A0P5J4L6 A0A0P5KX56 A0A0A1WWG5 A0A1B6HUG7 A0A0P5FSM7 A0A0N8CYY7 A0A1Y3AQ25 E9FQN6 A0A0P6CME8 W8BGG6 L7M528 A0A1A9Z7M9 A0A224YSK6 A0A1A9WBX2 A0A131YQF5 A0A1B0AP14 A0A1A9XVX4 A0A1A9VIC2 B4LKF7 A0A131XMB7 A0A091PWY7 A0A091QLX0 A0A1I7Q423 Q5ZK33 A0A091K1H7 E9GYB1 A0A226MWV8 A0A0P5LN53 A0A3Q1MMS8 G3U258 L8IIK8 Q0VCA3 F1N3R4 A0A2K6LZ68 A0A2K6QN82 K9INF8
PDB
3SKQ
E-value=1.17653e-30,
Score=335
Ontologies
GO
GO:0005739
GO:0016021
GO:0043022
GO:0005509
GO:0005743
GO:0006851
GO:0098793
GO:0071456
GO:0031966
GO:0005432
GO:0007269
GO:0070584
GO:0015386
GO:0015369
GO:0051562
GO:0099093
GO:0034214
GO:1900069
GO:0051260
GO:0042407
GO:0051560
GO:0005216
GO:0006811
GO:0016020
GO:0004970
GO:0005234
GO:0016787
GO:0004713
GO:0016491
GO:0015930
Topology
Subcellular location
Mitochondrion inner membrane
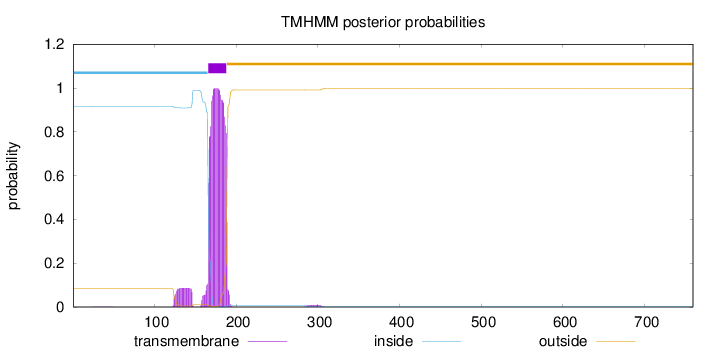
Length:
759
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.23285
Exp number, first 60 AAs:
0.01683
Total prob of N-in:
0.91630
inside
1 - 165
TMhelix
166 - 188
outside
189 - 759
Population Genetic Test Statistics
Pi
223.855773
Theta
184.296723
Tajima's D
0.936889
CLR
0.013018
CSRT
0.63756812159392
Interpretation
Uncertain
