Gene
KWMTBOMO13900
Pre Gene Modal
BGIBMGA011590
Annotation
PREDICTED:_uncharacterized_protein_LOC101738187_[Bombyx_mori]
Full name
Ionotropic receptor 25a
Location in the cell
PlasmaMembrane Reliability : 3.513
Sequence
CDS
ATGACGACGCAGAATATTAATGTTTTATTAATCAACGAAGAAAATAATGCCCTAGCTGAAAAATCCTTTGAAATAGCGAAAGAATATGTGAGACGTAATCCAAGTTTAGGATTAGCGATAGAACCAGTTATTGTCGTTGGAAACAGATCTGACGCCAAAACTTTCCTAGAAAATGTTTGCAGGAAATACAACGATATGCTATCATCGAAAAAAACACCTCACGTTGTCTTGGATTTCACAATGACCGGCGTCGGATCAGAAACTATAAAATCTTTCACCGCTGCATTGGCATTGCCAACGATATCGGGTTCATTCGGACAAACGGGAGATCTACGACAATGGAGGTCACTTAATGCTAACCAAACTAAATTTCTTCTGCAAGTGATGCCTCCTGCAGATATTCTACCTGAATCCATCAGAGCTATTGTTACTAAACAAGATATCACGAATGCCGCAATTATCTTTGACGAGCTTTTTGTTATGGACCATAAATACAAATCTTTACTACAAAATATTCCAACTCGTCATGTTATTACGCCAGTAAAAAGTTTTAACAAAGAAGACATCAAAACTCAATTACGCAGCCTGCGGGAATTGGATATAGTAAATTTCTTCATTGTCGGTAGTTTGAGGACTATCAAAAACGTGCTAGACGCAGCGGATGAAAATCAGTATTTCGGCCGGAAAACGGCCTGGTTCGCGTTCTCGTTAGATAAAGGAGACATTACTTGCGGCTGTAAAGATGCCACAATTGTTTATATGAGACCAACGCCAGACGCAAAGAGCAGAGATCGTTTGGGAAAAATCAAAACCACTTATAGCATGAACGGGGAGCCGGAAATTACATCTGCGTTCTACTTCGACTTGTCTTTGAGGACATTTTTGGCTGTCAAATCCCTACTGGACTCTGGCAAGTGGCCAAATAACATGAAATATATCACATGCGACGATTACGACGGCAAGAATACACCTAACAGGACCTTGGATCTTAAACTGGCTTTTCAAGAGGTGAAAGAGACTCCAACTTATGCTCCGTTTTATATCCCCGGAGACGACCCTATGAATGGAAGAAGCTACATGGAATTTAGTACTGATTTATCGGCAGTTACCGTCAAAGATGGTGCTTCTATAGGTAGTAAGGCCCTAGGAACTTGGAAAGCTGGTCTTAACAGTCCGCTGTCTTTGACTGATTCCGATAACATGAGCGATTATTCCGCTCAACTTGTGTACAGAGTTGTTACAGTCGAGCAACAACCTTTCATAATACGAGACGATAATGCCCCTAAAGGTTTTAAGGGATATTGCATTGACCTAATCGAGGAGATTCGTCAAATCGTTAAGTTTGATTACGAAGTGACTTTATCACCTGACGGTAATTTCGGAACGATGGACGAGAATGGTAACTGGAACGGCATCATAAAGGAATTGATCGAAAAGAGAGCGGATATCGCTCTAACTTCTCTGTCAGTAATGGCTGAGAGGGAAAATGTTGTAGATTTCACAGTGCCTTACTACGATTTGGTGGGTATAACGATAATGATGAAGTTGCCTAGAACACCGACGTCACTGTTTAAATTTTTAACAGTACTGGAAAATGACGTGTGGCTGTCGATACTTGCCGCTTACTTCTTTACCAGCTTTTTAATGTGGGTATTCGACAAATGGAGTCCGTATAGTTACCAGAACAATCGAGAAAAATACAAAGACGACGAGGAAAAACGCGAATTTACGCTTAAAGAATGTCTTTGGTTCTGTATGACGTCACTTACCCCGCAAGGCGGAGGGGAGGCGCCGAAAAACCTATCCGGGAGATTGCTTGCCGCTACTTGGTGGCTGTTCGGTTTTATAATAATAGCATCGTACACCGCGAATTTGGCGGCTTTCCTCACTGTATCTCGTTTGGATACTCCAATAGAATCGTTGGACGATCTATCGAAGCAATACAAAATACAGTACGCTCCATTGAATGGTTCTGCCGCCATGACATATTTCGAAAGGATGGCCGCTATCGAAGTCAGATTTTATGAAATATGGAAAGAAATGAGTTTGAACGACAGTCTAAGTGATGTCGAACGCGCTAAGCTAGCCGTATGGGATTATCCGGTCAGCGATAAATACAGTAAAATGTGGCAGGCCATGAAAGAGGCAGGCCTGCCTAACTCGATAGAGGAAGCCGTGCAAAGGGTGAGGGATTCGAAGAGCTCGAGCGAAGGTTTCGCCTGGCTGGGAGACGCTACGGACGTTAGGTACTATGTCCTGACGAGCTGCGATCTTCAGATGGTCGGAGACGAATTCTCGAGGAAGCCTTACGCTATTGCCGTGCAGCAAGGATCTCCTTTGAAAGACCAGTTTAACAACGCGATTCTGCAGCTACTTAACAGACGTAGACTAGAGAAGCTCAAAGAAAACTGGTGGAACAATAATCCCAAAGCGATGAAATGTGAAAAGCAAGATGACCAGTCCGACGGGATCTCTATTCAGAACATCGGAGGTGTTTTCATAGTTATCTTTATGGGGATAGGACTTGCTTGCATCACTCTGGGCGTCGAGTACTGGTGGTATAAATGGAGGCGACGTCCAATTGTCGGTGACGTCACACAGGTGGAACCAGCTAAATCAACGAGAAATAACATCGGAAATTTCGTCAAAGGCGAAGGATTCACTTTTCGTTCTAGAAACTTCGGTCTCTCCGATTTAAAGCAGAAATTTTAA
Protein
MTTQNINVLLINEENNALAEKSFEIAKEYVRRNPSLGLAIEPVIVVGNRSDAKTFLENVCRKYNDMLSSKKTPHVVLDFTMTGVGSETIKSFTAALALPTISGSFGQTGDLRQWRSLNANQTKFLLQVMPPADILPESIRAIVTKQDITNAAIIFDELFVMDHKYKSLLQNIPTRHVITPVKSFNKEDIKTQLRSLRELDIVNFFIVGSLRTIKNVLDAADENQYFGRKTAWFAFSLDKGDITCGCKDATIVYMRPTPDAKSRDRLGKIKTTYSMNGEPEITSAFYFDLSLRTFLAVKSLLDSGKWPNNMKYITCDDYDGKNTPNRTLDLKLAFQEVKETPTYAPFYIPGDDPMNGRSYMEFSTDLSAVTVKDGASIGSKALGTWKAGLNSPLSLTDSDNMSDYSAQLVYRVVTVEQQPFIIRDDNAPKGFKGYCIDLIEEIRQIVKFDYEVTLSPDGNFGTMDENGNWNGIIKELIEKRADIALTSLSVMAERENVVDFTVPYYDLVGITIMMKLPRTPTSLFKFLTVLENDVWLSILAAYFFTSFLMWVFDKWSPYSYQNNREKYKDDEEKREFTLKECLWFCMTSLTPQGGGEAPKNLSGRLLAATWWLFGFIIIASYTANLAAFLTVSRLDTPIESLDDLSKQYKIQYAPLNGSAAMTYFERMAAIEVRFYEIWKEMSLNDSLSDVERAKLAVWDYPVSDKYSKMWQAMKEAGLPNSIEEAVQRVRDSKSSSEGFAWLGDATDVRYYVLTSCDLQMVGDEFSRKPYAIAVQQGSPLKDQFNNAILQLLNRRRLEKLKENWWNNNPKAMKCEKQDDQSDGISIQNIGGVFIVIFMGIGLACITLGVEYWWYKWRRRPIVGDVTQVEPAKSTRNNIGNFVKGEGFTFRSRNFGLSDLKQKF
Summary
Description
Integral part of various neural sensory systems in the antenna that provide the neural basis for the response to environmental changes in temperature (thermosensation), humidity (hygrosensation) and odor detection (PubMed:21220098, PubMed:27161501, PubMed:27656904). Required for odor-evoked electrophysiological responses in multiple neuron classes in the antenna and is likely to function as part of an olfactory receptor complex with Ir76a and Ir76b (PubMed:21220098). Together with Ir21a and Ir93a, mediates the response of the larval dorsal organ cool cells, a trio of cool-responsive neurons, to cooling and is required for cool avoidance behavior (PubMed:27126188, PubMed:27161501, PubMed:27656904). Required in chordonotal organ neurons for behavioral synchronization to low-amplitude temperature cycles and mediates circadian clock resetting by temperature (PubMed:26580016). Together with Ir40a and Ir93a, mediates the response of the hydrosensory sacculus neurons to changes in relative humidity, and is required for dry detection and humidiy preference behavior (PubMed:27161501, PubMed:27656904).
Subunit
Interacts with nocte.
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Keywords
Behavior
Biological rhythms
Cell membrane
Cell projection
Complete proteome
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Olfaction
Receptor
Reference proteome
Sensory transduction
Signal
Transmembrane
Transmembrane helix
Transport
Feature
chain Ionotropic receptor 25a
Uniprot
H9JPY5
A0A0K8TUE0
A0A1X9PBX1
A0A3Q9NC15
A0A3G6V6N5
A0A0F7QIF9
+ More
A0A345BF42 A0A1Q1PP75 H9A5R7 A0A223HCW8 A0A223HDA2 A0A1B3P5G0 A0A3S2L9B5 A0A194PKX2 A0A212FLC1 A0A0B5A1Z1 A0A2A4K2Y9 A0A140G9H8 A0A1V1WBQ6 A0A076E7N8 A0A1B3B709 A0A076E7Q0 A0A1P8SKC2 A0A2K8GKW5 A0A2H1VEZ6 A0A0L7LS05 A0A0C5D6R2 D6WAZ2 A0A2P9JYA8 A0A182H4U4 Q16US4 A0A1S4FNE0 A0A182QUT1 A0A182N8W0 A0A2H8TLX6 J9K5M8 A0A385IUV0 A0A2S2QQE6 A0A182RAE4 A0A385IUV1 A0A182I0W0 Q7PMF1 A0A182M9H0 A0A182U1Q4 A0A182USW2 A0A182X1S1 A0A182IST6 A0A182VST0 A0A182PPU8 A0A182JSA2 A0A0S3J2L0 A0A084W5L5 A0A1B0CCC1 A0A310SBI5 A0A2P1JHH0 A0A2S2NU70 A0A1J1HGH0 E0VBE7 A0A182KJX4 B4JQB5 W8BJ37 B4MDP5 A0A348AZW9 A0A034WU95 B4KGP7 A0A0K8VV08 B4N0H8 A0A182YN05 A0A0X8DBS6 A0A146KXM4 A0A1B3B7C9 B3MLG4 B4G8X6 E9NA96 C6TPA2 Q9VR32 M9PC56 Q29MQ4 A0A1I8PNS9 A0A0J9TFM2 A0A0J9QWN2 B4NZ14 B4Q332 A0A3B0JB79 B4I3D0 R9PSM2 U4U5U6 A0A1W4UW16 J7H936 A0A0M3QT25 A0A0L0CB94 A0A068F4U6 B3N4A2 A0A067QT16 T1H988 A0A2I4PH03 N6T537 A0A1I8NTP0 A0A1A9XFQ3 A0A1A9UPZ5
A0A345BF42 A0A1Q1PP75 H9A5R7 A0A223HCW8 A0A223HDA2 A0A1B3P5G0 A0A3S2L9B5 A0A194PKX2 A0A212FLC1 A0A0B5A1Z1 A0A2A4K2Y9 A0A140G9H8 A0A1V1WBQ6 A0A076E7N8 A0A1B3B709 A0A076E7Q0 A0A1P8SKC2 A0A2K8GKW5 A0A2H1VEZ6 A0A0L7LS05 A0A0C5D6R2 D6WAZ2 A0A2P9JYA8 A0A182H4U4 Q16US4 A0A1S4FNE0 A0A182QUT1 A0A182N8W0 A0A2H8TLX6 J9K5M8 A0A385IUV0 A0A2S2QQE6 A0A182RAE4 A0A385IUV1 A0A182I0W0 Q7PMF1 A0A182M9H0 A0A182U1Q4 A0A182USW2 A0A182X1S1 A0A182IST6 A0A182VST0 A0A182PPU8 A0A182JSA2 A0A0S3J2L0 A0A084W5L5 A0A1B0CCC1 A0A310SBI5 A0A2P1JHH0 A0A2S2NU70 A0A1J1HGH0 E0VBE7 A0A182KJX4 B4JQB5 W8BJ37 B4MDP5 A0A348AZW9 A0A034WU95 B4KGP7 A0A0K8VV08 B4N0H8 A0A182YN05 A0A0X8DBS6 A0A146KXM4 A0A1B3B7C9 B3MLG4 B4G8X6 E9NA96 C6TPA2 Q9VR32 M9PC56 Q29MQ4 A0A1I8PNS9 A0A0J9TFM2 A0A0J9QWN2 B4NZ14 B4Q332 A0A3B0JB79 B4I3D0 R9PSM2 U4U5U6 A0A1W4UW16 J7H936 A0A0M3QT25 A0A0L0CB94 A0A068F4U6 B3N4A2 A0A067QT16 T1H988 A0A2I4PH03 N6T537 A0A1I8NTP0 A0A1A9XFQ3 A0A1A9UPZ5
Pubmed
19121390
26017144
30465940
25803580
29727827
28150741
+ More
22363688 27006164 27538507 26354079 22118469 27004525 24998398 28736530 26227816 25665775 18362917 19820115 26483478 17510324 30195211 12364791 26626891 24438588 20566863 20966253 17994087 24495485 30026095 25348373 25244985 29403074 26823975 27171401 21220098 10731132 12537572 19135896 26580016 27126188 27161501 27656904 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 23517120 23537049 25315136 26108605 25880816 24845553
22363688 27006164 27538507 26354079 22118469 27004525 24998398 28736530 26227816 25665775 18362917 19820115 26483478 17510324 30195211 12364791 26626891 24438588 20566863 20966253 17994087 24495485 30026095 25348373 25244985 29403074 26823975 27171401 21220098 10731132 12537572 19135896 26580016 27126188 27161501 27656904 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 23517120 23537049 25315136 26108605 25880816 24845553
EMBL
BABH01015551
BABH01015552
GCVX01000133
JAI18097.1
KX084508
ARO76463.1
+ More
MH050691 AZT78940.1 MH193958 AZB49404.1 LC017784 BAR64798.1 MG820695 AXF48866.1 KY325450 AQM73611.1 JN836717 AFC91757.2 KY283568 AST36228.1 KY283699 AST36358.1 KX655898 AOG12847.1 RSAL01000081 RVE48574.1 KQ459601 KPI94081.1 AGBW02007799 OWR54543.1 KJ542744 AJD81628.1 NWSH01000246 PCG78050.1 KU702630 AMM70657.1 GENK01000125 JAV45788.1 KF487714 AII01112.1 KT588089 AOE47999.1 KF487724 AII01122.1 KX252767 APY22696.1 KX585393 ARO70286.1 ODYU01002206 SOQ39397.1 JTDY01000213 KOB78263.1 KP296780 AJO62244.1 KQ971311 EEZ98989.2 KY817099 AVH87315.1 JXUM01110311 JXUM01110312 JXUM01110313 JXUM01110314 JXUM01110315 KQ565401 KXJ71028.1 CH477615 EAT38264.1 AXCN02001867 GFXV01003146 MBW14951.1 ABLF02036596 MH230261 AXY87920.1 GGMS01010745 MBY79948.1 MH230262 AXY87921.1 APCN01005276 AAAB01008980 EAA13931.5 AXCM01000163 KT381529 ALR72535.1 ATLV01020673 KE525304 KFB45509.1 AJWK01006611 GGOB01000051 MCH29447.1 MG766215 AVN97885.1 GGMR01008132 MBY20751.1 CVRI01000001 CRK86500.1 DS235028 EEB10703.1 CH916372 EDV99095.1 GAMC01005235 JAC01321.1 CH940661 EDW71306.2 FX985960 BBD43495.1 GAKP01001202 JAC57750.1 CH933807 EDW13247.2 GDHF01009934 JAI42380.1 CH963920 EDW77591.2 KT381711 PYGN01001004 AMA98202.1 PSN38557.1 GDHC01017435 JAQ01194.1 KU291867 AOE48117.1 CH902620 EDV31713.2 CH479180 EDW28806.1 HQ600588 AE014134 ADU79032.1 BT099588 ACU43550.1 AAF50976.2 AGB92587.1 AGB92586.1 CH379060 EAL33639.3 CM002910 KMY88125.1 KMY88124.1 CM000157 EDW87678.2 CM000361 EDX03748.1 OUUW01000004 SPP79275.1 CH480820 EDW54275.1 GABX01000048 JAA74471.1 KB632161 ERL89264.1 JX263304 AFP89966.1 CP012523 ALC38173.1 JRES01000753 KNC28739.1 KJ702119 AID61273.1 CH954177 EDV57772.2 KK853313 KDR08634.1 ACPB03014189 KU523597 APZ81419.1 APGK01043536 APGK01043537 KB741015 ENN75289.1
MH050691 AZT78940.1 MH193958 AZB49404.1 LC017784 BAR64798.1 MG820695 AXF48866.1 KY325450 AQM73611.1 JN836717 AFC91757.2 KY283568 AST36228.1 KY283699 AST36358.1 KX655898 AOG12847.1 RSAL01000081 RVE48574.1 KQ459601 KPI94081.1 AGBW02007799 OWR54543.1 KJ542744 AJD81628.1 NWSH01000246 PCG78050.1 KU702630 AMM70657.1 GENK01000125 JAV45788.1 KF487714 AII01112.1 KT588089 AOE47999.1 KF487724 AII01122.1 KX252767 APY22696.1 KX585393 ARO70286.1 ODYU01002206 SOQ39397.1 JTDY01000213 KOB78263.1 KP296780 AJO62244.1 KQ971311 EEZ98989.2 KY817099 AVH87315.1 JXUM01110311 JXUM01110312 JXUM01110313 JXUM01110314 JXUM01110315 KQ565401 KXJ71028.1 CH477615 EAT38264.1 AXCN02001867 GFXV01003146 MBW14951.1 ABLF02036596 MH230261 AXY87920.1 GGMS01010745 MBY79948.1 MH230262 AXY87921.1 APCN01005276 AAAB01008980 EAA13931.5 AXCM01000163 KT381529 ALR72535.1 ATLV01020673 KE525304 KFB45509.1 AJWK01006611 GGOB01000051 MCH29447.1 MG766215 AVN97885.1 GGMR01008132 MBY20751.1 CVRI01000001 CRK86500.1 DS235028 EEB10703.1 CH916372 EDV99095.1 GAMC01005235 JAC01321.1 CH940661 EDW71306.2 FX985960 BBD43495.1 GAKP01001202 JAC57750.1 CH933807 EDW13247.2 GDHF01009934 JAI42380.1 CH963920 EDW77591.2 KT381711 PYGN01001004 AMA98202.1 PSN38557.1 GDHC01017435 JAQ01194.1 KU291867 AOE48117.1 CH902620 EDV31713.2 CH479180 EDW28806.1 HQ600588 AE014134 ADU79032.1 BT099588 ACU43550.1 AAF50976.2 AGB92587.1 AGB92586.1 CH379060 EAL33639.3 CM002910 KMY88125.1 KMY88124.1 CM000157 EDW87678.2 CM000361 EDX03748.1 OUUW01000004 SPP79275.1 CH480820 EDW54275.1 GABX01000048 JAA74471.1 KB632161 ERL89264.1 JX263304 AFP89966.1 CP012523 ALC38173.1 JRES01000753 KNC28739.1 KJ702119 AID61273.1 CH954177 EDV57772.2 KK853313 KDR08634.1 ACPB03014189 KU523597 APZ81419.1 APGK01043536 APGK01043537 KB741015 ENN75289.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000007266 UP000069940 UP000249989 UP000008820 UP000075886 UP000075884 UP000007819 UP000075900 UP000075840 UP000007062 UP000075883 UP000075902 UP000075903 UP000076407 UP000075880 UP000075920 UP000075885 UP000075881 UP000030765 UP000092461 UP000183832 UP000009046 UP000075882 UP000001070 UP000008792 UP000009192 UP000007798 UP000076408 UP000245037 UP000007801 UP000008744 UP000000803 UP000001819 UP000095300 UP000002282 UP000000304 UP000268350 UP000001292 UP000030742 UP000192221 UP000095301 UP000092553 UP000037069 UP000008711 UP000027135 UP000015103 UP000019118 UP000092443 UP000078200
UP000007266 UP000069940 UP000249989 UP000008820 UP000075886 UP000075884 UP000007819 UP000075900 UP000075840 UP000007062 UP000075883 UP000075902 UP000075903 UP000076407 UP000075880 UP000075920 UP000075885 UP000075881 UP000030765 UP000092461 UP000183832 UP000009046 UP000075882 UP000001070 UP000008792 UP000009192 UP000007798 UP000076408 UP000245037 UP000007801 UP000008744 UP000000803 UP000001819 UP000095300 UP000002282 UP000000304 UP000268350 UP000001292 UP000030742 UP000192221 UP000095301 UP000092553 UP000037069 UP000008711 UP000027135 UP000015103 UP000019118 UP000092443 UP000078200
Interpro
ProteinModelPortal
H9JPY5
A0A0K8TUE0
A0A1X9PBX1
A0A3Q9NC15
A0A3G6V6N5
A0A0F7QIF9
+ More
A0A345BF42 A0A1Q1PP75 H9A5R7 A0A223HCW8 A0A223HDA2 A0A1B3P5G0 A0A3S2L9B5 A0A194PKX2 A0A212FLC1 A0A0B5A1Z1 A0A2A4K2Y9 A0A140G9H8 A0A1V1WBQ6 A0A076E7N8 A0A1B3B709 A0A076E7Q0 A0A1P8SKC2 A0A2K8GKW5 A0A2H1VEZ6 A0A0L7LS05 A0A0C5D6R2 D6WAZ2 A0A2P9JYA8 A0A182H4U4 Q16US4 A0A1S4FNE0 A0A182QUT1 A0A182N8W0 A0A2H8TLX6 J9K5M8 A0A385IUV0 A0A2S2QQE6 A0A182RAE4 A0A385IUV1 A0A182I0W0 Q7PMF1 A0A182M9H0 A0A182U1Q4 A0A182USW2 A0A182X1S1 A0A182IST6 A0A182VST0 A0A182PPU8 A0A182JSA2 A0A0S3J2L0 A0A084W5L5 A0A1B0CCC1 A0A310SBI5 A0A2P1JHH0 A0A2S2NU70 A0A1J1HGH0 E0VBE7 A0A182KJX4 B4JQB5 W8BJ37 B4MDP5 A0A348AZW9 A0A034WU95 B4KGP7 A0A0K8VV08 B4N0H8 A0A182YN05 A0A0X8DBS6 A0A146KXM4 A0A1B3B7C9 B3MLG4 B4G8X6 E9NA96 C6TPA2 Q9VR32 M9PC56 Q29MQ4 A0A1I8PNS9 A0A0J9TFM2 A0A0J9QWN2 B4NZ14 B4Q332 A0A3B0JB79 B4I3D0 R9PSM2 U4U5U6 A0A1W4UW16 J7H936 A0A0M3QT25 A0A0L0CB94 A0A068F4U6 B3N4A2 A0A067QT16 T1H988 A0A2I4PH03 N6T537 A0A1I8NTP0 A0A1A9XFQ3 A0A1A9UPZ5
A0A345BF42 A0A1Q1PP75 H9A5R7 A0A223HCW8 A0A223HDA2 A0A1B3P5G0 A0A3S2L9B5 A0A194PKX2 A0A212FLC1 A0A0B5A1Z1 A0A2A4K2Y9 A0A140G9H8 A0A1V1WBQ6 A0A076E7N8 A0A1B3B709 A0A076E7Q0 A0A1P8SKC2 A0A2K8GKW5 A0A2H1VEZ6 A0A0L7LS05 A0A0C5D6R2 D6WAZ2 A0A2P9JYA8 A0A182H4U4 Q16US4 A0A1S4FNE0 A0A182QUT1 A0A182N8W0 A0A2H8TLX6 J9K5M8 A0A385IUV0 A0A2S2QQE6 A0A182RAE4 A0A385IUV1 A0A182I0W0 Q7PMF1 A0A182M9H0 A0A182U1Q4 A0A182USW2 A0A182X1S1 A0A182IST6 A0A182VST0 A0A182PPU8 A0A182JSA2 A0A0S3J2L0 A0A084W5L5 A0A1B0CCC1 A0A310SBI5 A0A2P1JHH0 A0A2S2NU70 A0A1J1HGH0 E0VBE7 A0A182KJX4 B4JQB5 W8BJ37 B4MDP5 A0A348AZW9 A0A034WU95 B4KGP7 A0A0K8VV08 B4N0H8 A0A182YN05 A0A0X8DBS6 A0A146KXM4 A0A1B3B7C9 B3MLG4 B4G8X6 E9NA96 C6TPA2 Q9VR32 M9PC56 Q29MQ4 A0A1I8PNS9 A0A0J9TFM2 A0A0J9QWN2 B4NZ14 B4Q332 A0A3B0JB79 B4I3D0 R9PSM2 U4U5U6 A0A1W4UW16 J7H936 A0A0M3QT25 A0A0L0CB94 A0A068F4U6 B3N4A2 A0A067QT16 T1H988 A0A2I4PH03 N6T537 A0A1I8NTP0 A0A1A9XFQ3 A0A1A9UPZ5
PDB
5KUF
E-value=2.84655e-67,
Score=652
Ontologies
GO
GO:0016021
GO:0004970
GO:0005886
GO:0006811
GO:0032281
GO:0004971
GO:0016747
GO:0045211
GO:0030054
GO:0010378
GO:0015026
GO:0009649
GO:0071683
GO:0004984
GO:0019226
GO:0005929
GO:0007610
GO:0050907
GO:0043204
GO:0030424
GO:0050911
GO:0048511
GO:0015276
GO:0004872
GO:0005216
GO:0016020
GO:0005234
GO:0016787
GO:0004713
GO:0016491
GO:0015930
GO:0038023
Topology
Subcellular location
Cell membrane Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
Cell projection Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
Axon Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
Dendrite Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
Perikaryon Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
Cilium Low levels detected in the axon segment adjacent to the perikaryon in some sensory neurons of the antenna but not detected along axons as they enter the brain or at synapses within antennal lobe glomeruli. In coeloconic neurons, prominently expressed both in the perikaryon and in the distal tip of the dendrite which corresponds to the ciliated outer dendritic segment innervating the sensory hair. Relatively low levels detected in inner dendrites (PubMed:19135896). Detected in dendritic bulbs of the dorsal organ cool cells (PubMed:27126188). With evidence from 4 publications.
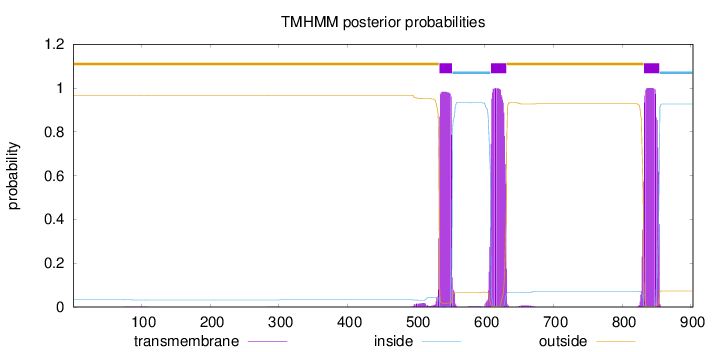
Length:
903
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
64.90696
Exp number, first 60 AAs:
0.00129
Total prob of N-in:
0.03315
outside
1 - 533
TMhelix
534 - 552
inside
553 - 608
TMhelix
609 - 631
outside
632 - 831
TMhelix
832 - 854
inside
855 - 903
Population Genetic Test Statistics
Pi
283.942141
Theta
186.501098
Tajima's D
0.339548
CLR
0.648541
CSRT
0.468126593670317
Interpretation
Uncertain
