Pre Gene Modal
BGIBMGA011429
Annotation
cAMP-dependent_protein_kinase_C1_[Bombyx_mori]
Full name
cAMP-dependent protein kinase catalytic subunit 1
Alternative Name
Protein kinase DC0
Location in the cell
Cytoplasmic Reliability : 3.687
Sequence
CDS
ATGGGCAACAATGCTGCCACCGCTAATAAAAAGGTGGACGCCGCCGAAAGCGTCAAAGAATTCCTCGACCAGGCCAAAGAGGACTTCGAGGAGAAATGGAAGAAGAATCCTACCAACACCGCCGGACTGAACGATTTCGAACGGATAAAAACCCTAGGTACTGGCTCGTTTGGACGAGTCATGATAGTTCAACATAAACCTACTAAAGAATATTACGCTATGAAAATATTAGATAAACAGAAAGTGGTTAAATTGAAACAGGTGGAGCACACTCTAAACGAGAAACGTATCTTACAAGCCATCAACTTTCCGTTTTTGGTGAGCCTTAAATTCCATTTCAAGGACAACTCTAACTTGTACATGGTGTTGGAGTATGTGCCTGGAGGGGAAATGTTCTCGCATTTGCGCAAGGTGGGTCGGTTCTCGGAGCCGCATTCGCGATTCTACGCAGCTCAAATAGTCTTAGCATTTGAGTACCTTCACTATTTGGACCTTATTTATCGGGATCTGAAACCTGAAAACTTGCTAATTGATTCTCAGGGTTACTTAAAAGTTACAGATTTCGGATTCGCTAAACGTGTGAAAGGCCGCACGTGGACTTTGTGCGGCACTCCTGAGTATCTTGCACCTGAAATCATTCTATCCAAGGGATACAACAAAGCTGTGGATTGGTGGGCGTTAGGAGTGCTCGTGTATGAGATGGCGGCTGGCTACCCGCCCTTTTTCGCTGATCAACCCATTCAGATCTATGAAAAAATTGTGTCTGGCAAAGTACGATTTCCATCACATTTTGGTTCAGATCTGAAGGATCTGCTTCGTAACCTACTTCAAGTAGATTTGACCAAACGCTATGGTAACTTGAAGGCTGGTGTCAATGATATAAAAGGTCATAAATGGTTTGCCACCACCGACTGGATTGCCGTCTTCCAGAAGAAGATAGAGGCTCCATTCATACCTCGCTGCAAAGGACCCGGTGACACAAGCAATTTTGATGATTATGAAGAAGAAGCTCTTCGTATTTCTTCCACTGAAAAGTGTGCAAAGGAATTTGCAGAGTTCTGA
Protein
MGNNAATANKKVDAAESVKEFLDQAKEDFEEKWKKNPTNTAGLNDFERIKTLGTGSFGRVMIVQHKPTKEYYAMKILDKQKVVKLKQVEHTLNEKRILQAINFPFLVSLKFHFKDNSNLYMVLEYVPGGEMFSHLRKVGRFSEPHSRFYAAQIVLAFEYLHYLDLIYRDLKPENLLIDSQGYLKVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLVYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFGSDLKDLLRNLLQVDLTKRYGNLKAGVNDIKGHKWFATTDWIAVFQKKIEAPFIPRCKGPGDTSNFDDYEEEALRISSTEKCAKEFAEF
Summary
Description
Serine/threonine-protein kinase involved in memory formation (PubMed:29473541). Promotes long-term memory by phosphorylating meng and by regulating CrebB protein stability and activity (PubMed:29473541). As part of ethanol response in the glia, mediates ethanol-induced structural remodeling of actin cytoskeleton and perineurial membrane topology when anchored to the membrane (PubMed:29444420).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Composed of two regulatory chains and two catalytic chains.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cAMP subfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cAMP subfamily.
Keywords
ATP-binding
cAMP
Complete proteome
Kinase
Lipoprotein
Myristate
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain cAMP-dependent protein kinase catalytic subunit 1
Uniprot
A5JNM1
A0A0S2VHW6
A0A2A4K291
A0A194RME0
A0A212FLD3
A0A194PMX1
+ More
A0A0L7L866 A0A1W4WWQ6 D6WC07 A0A1Y1MUE8 N6UW64 A0A1L8DM17 A0A0K8TSM1 A0A1L8DKK8 A0A1L8DLK2 T1DEF5 A0A1Q3FP44 Q1HQW4 A0A023EQD8 A0A0C9RJF4 B0XFG0 B4HW89 A0A1Y9IZU8 A0A3F2YVZ5 U5EXF6 A0A182LYM3 A0A182SXB9 A0A182VLF0 A0A182TV87 A0A182L915 Q7PXF7 A0A1A9XGX3 T1E804 A0A2M3ZG62 A0A2M4A7I4 W5JN41 A0A182JM51 A0A1A9UCN4 B4G7Y8 A0A1B0A4X0 A0A1B0C119 A0A1W4W8R0 B4M9H6 B4KH13 Q29NB1 B4NY66 B4MVA0 B4JCE3 B3N8G6 B4Q7V0 B3MUX5 A0A026WR62 P12370 A0A0G4AJS6 W8C0H4 A0A195EJF1 A0A2R7VYK8 F4X2B5 A0A195BCI3 A0A0J7KSG5 A0A195FN55 A0A158NJP0 A0A151X6H1 T1PGE9 A0A1I8NM79 E9IFT9 A0A2A3EBH8 E2BPC9 A0A2H8TN08 J9JK31 A0A2S2NEM0 A0A2S2PZY9 Q9NAS6 V9IMJ8 A0A087ZX62 A0A0A1X7C9 A0A0L7R929 A0A0C9RGE4 A0A0K8V0C2 A0A154P8G8 E0VMS0 A0A224XR64 A0A023F5V8 A0A0M9A1X7 A0A2J7PIG0 T1IA19 A0A0A9ZCM8 A0A067RHU0 A0A1B6LQI5 A0A1B6GP45 A0A1S3DFC7 A0A034VJA2 A0A182QBI4 E9GNI4 A0A1S3JH22 T1E265 A0A1S3JGA5 A0A0P6AK16 A0A076FKS1 A0A0P6DJB0
A0A0L7L866 A0A1W4WWQ6 D6WC07 A0A1Y1MUE8 N6UW64 A0A1L8DM17 A0A0K8TSM1 A0A1L8DKK8 A0A1L8DLK2 T1DEF5 A0A1Q3FP44 Q1HQW4 A0A023EQD8 A0A0C9RJF4 B0XFG0 B4HW89 A0A1Y9IZU8 A0A3F2YVZ5 U5EXF6 A0A182LYM3 A0A182SXB9 A0A182VLF0 A0A182TV87 A0A182L915 Q7PXF7 A0A1A9XGX3 T1E804 A0A2M3ZG62 A0A2M4A7I4 W5JN41 A0A182JM51 A0A1A9UCN4 B4G7Y8 A0A1B0A4X0 A0A1B0C119 A0A1W4W8R0 B4M9H6 B4KH13 Q29NB1 B4NY66 B4MVA0 B4JCE3 B3N8G6 B4Q7V0 B3MUX5 A0A026WR62 P12370 A0A0G4AJS6 W8C0H4 A0A195EJF1 A0A2R7VYK8 F4X2B5 A0A195BCI3 A0A0J7KSG5 A0A195FN55 A0A158NJP0 A0A151X6H1 T1PGE9 A0A1I8NM79 E9IFT9 A0A2A3EBH8 E2BPC9 A0A2H8TN08 J9JK31 A0A2S2NEM0 A0A2S2PZY9 Q9NAS6 V9IMJ8 A0A087ZX62 A0A0A1X7C9 A0A0L7R929 A0A0C9RGE4 A0A0K8V0C2 A0A154P8G8 E0VMS0 A0A224XR64 A0A023F5V8 A0A0M9A1X7 A0A2J7PIG0 T1IA19 A0A0A9ZCM8 A0A067RHU0 A0A1B6LQI5 A0A1B6GP45 A0A1S3DFC7 A0A034VJA2 A0A182QBI4 E9GNI4 A0A1S3JH22 T1E265 A0A1S3JGA5 A0A0P6AK16 A0A076FKS1 A0A0P6DJB0
EC Number
2.7.11.11
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
28004739 23537049 26369729 24330624 17204158 17510324 24945155 26483478 17994087 20966253 12364791 14747013 17210077 20920257 23761445 15632085 17550304 22936249 18057021 24508170 30249741 2828348 3215511 10731132 12537572 12537569 29444420 29473541 26252388 24495485 21719571 21347285 25315136 21282665 20798317 11422513 25830018 20566863 25474469 25401762 26823975 24845553 25348373 21292972
28004739 23537049 26369729 24330624 17204158 17510324 24945155 26483478 17994087 20966253 12364791 14747013 17210077 20920257 23761445 15632085 17550304 22936249 18057021 24508170 30249741 2828348 3215511 10731132 12537572 12537569 29444420 29473541 26252388 24495485 21719571 21347285 25315136 21282665 20798317 11422513 25830018 20566863 25474469 25401762 26823975 24845553 25348373 21292972
EMBL
BABH01015550
EF551340
ABQ41111.1
KT207930
ALP86605.1
NWSH01000246
+ More
PCG78048.1 KQ460205 KPJ17156.1 AGBW02007798 OWR54554.1 KQ459601 KPI94079.1 JTDY01002374 KOB71550.1 KQ971309 EEZ99369.1 GEZM01025015 JAV87646.1 APGK01006540 APGK01007788 APGK01021688 KB740353 KB737454 KB736818 KB632345 ENN80824.1 ENN83018.1 ENN83127.1 ERL93117.1 GFDF01006704 JAV07380.1 GDAI01000658 JAI16945.1 GFDF01007169 JAV06915.1 GFDF01006746 JAV07338.1 GALA01001042 JAA93810.1 GFDL01005792 JAV29253.1 DQ440330 CH477432 ABF18363.1 EAT41043.1 EAT41044.1 EAT41045.1 JXUM01089160 GAPW01002290 KQ563750 JAC11308.1 KXJ73385.1 GBYB01008335 JAG78102.1 DS232922 EDS26798.1 CH480818 EDW52284.1 GANO01000978 JAB58893.1 AXCM01000734 AAAB01008987 EAA01109.4 GAMD01003173 JAA98417.1 GGFM01006732 MBW27483.1 GGFK01003432 MBW36753.1 ADMH02000631 GGFL01004203 ETN65541.1 MBW68381.1 AXCP01003648 CH479180 EDW28486.1 JXJN01023868 CH940654 EDW57852.2 CH933807 EDW12224.2 CH379060 EAL33431.3 CM000157 EDW88668.2 CH963857 EDW76445.2 CH916368 EDW03097.1 CH954177 EDV58389.2 CM000361 CM002910 EDX04382.1 KMY89299.1 CH902624 EDV33040.2 KPU74269.1 KK107144 QOIP01000003 EZA57589.1 RLU24862.1 M18655 X16969 AE014134 AY069425 KR133400 AKM70870.1 GAMC01004015 GAMC01004014 GAMC01004013 GAMC01004012 JAC02544.1 KQ978782 KYN28380.1 KK854174 PTY12587.1 GL888575 EGI59411.1 KQ976526 KYM81932.1 LBMM01003676 KMQ93256.1 KQ981424 KYN41831.1 ADTU01018100 KQ982482 KYQ55934.1 KA647769 AFP62398.1 GL762903 EFZ20550.1 KZ288292 PBC29085.1 GL449613 EFN82454.1 GFXV01003710 MBW15515.1 ABLF02039909 GGMR01002966 MBY15585.1 GGMS01001883 MBY71086.1 AJ271674 CAC00652.1 JR053189 AEY61871.1 GBXI01012766 GBXI01007088 JAD01526.1 JAD07204.1 KQ414628 KOC67375.1 GBYB01006111 JAG75878.1 GDHF01025164 GDHF01022334 GDHF01021782 GDHF01020628 GDHF01019942 GDHF01013839 GDHF01009740 GDHF01007804 GDHF01006993 GDHF01006140 GDHF01004604 JAI27150.1 JAI29980.1 JAI30532.1 JAI31686.1 JAI32372.1 JAI38475.1 JAI42574.1 JAI44510.1 JAI45321.1 JAI46174.1 JAI47710.1 KQ434844 KZC08162.1 AAZO01003698 DS235324 EEB14676.1 GFTR01005496 JAW10930.1 GBBI01002143 JAC16569.1 KQ435794 KOX74139.1 NEVH01025127 PNF16136.1 ACPB03017770 GBHO01004044 GBRD01009005 GDHC01004987 JAG39560.1 JAG56816.1 JAQ13642.1 KK852463 KDR23367.1 GEBQ01022333 GEBQ01014002 JAT17644.1 JAT25975.1 GECZ01005582 GECZ01000688 JAS64187.1 JAS69081.1 GAKP01016379 GAKP01016378 JAC42574.1 AXCN02000029 GL732555 EFX78769.1 GALA01001014 JAA93838.1 GDIP01028209 JAM75506.1 KF516617 AII16521.1 GDIQ01085762 JAN08975.1
PCG78048.1 KQ460205 KPJ17156.1 AGBW02007798 OWR54554.1 KQ459601 KPI94079.1 JTDY01002374 KOB71550.1 KQ971309 EEZ99369.1 GEZM01025015 JAV87646.1 APGK01006540 APGK01007788 APGK01021688 KB740353 KB737454 KB736818 KB632345 ENN80824.1 ENN83018.1 ENN83127.1 ERL93117.1 GFDF01006704 JAV07380.1 GDAI01000658 JAI16945.1 GFDF01007169 JAV06915.1 GFDF01006746 JAV07338.1 GALA01001042 JAA93810.1 GFDL01005792 JAV29253.1 DQ440330 CH477432 ABF18363.1 EAT41043.1 EAT41044.1 EAT41045.1 JXUM01089160 GAPW01002290 KQ563750 JAC11308.1 KXJ73385.1 GBYB01008335 JAG78102.1 DS232922 EDS26798.1 CH480818 EDW52284.1 GANO01000978 JAB58893.1 AXCM01000734 AAAB01008987 EAA01109.4 GAMD01003173 JAA98417.1 GGFM01006732 MBW27483.1 GGFK01003432 MBW36753.1 ADMH02000631 GGFL01004203 ETN65541.1 MBW68381.1 AXCP01003648 CH479180 EDW28486.1 JXJN01023868 CH940654 EDW57852.2 CH933807 EDW12224.2 CH379060 EAL33431.3 CM000157 EDW88668.2 CH963857 EDW76445.2 CH916368 EDW03097.1 CH954177 EDV58389.2 CM000361 CM002910 EDX04382.1 KMY89299.1 CH902624 EDV33040.2 KPU74269.1 KK107144 QOIP01000003 EZA57589.1 RLU24862.1 M18655 X16969 AE014134 AY069425 KR133400 AKM70870.1 GAMC01004015 GAMC01004014 GAMC01004013 GAMC01004012 JAC02544.1 KQ978782 KYN28380.1 KK854174 PTY12587.1 GL888575 EGI59411.1 KQ976526 KYM81932.1 LBMM01003676 KMQ93256.1 KQ981424 KYN41831.1 ADTU01018100 KQ982482 KYQ55934.1 KA647769 AFP62398.1 GL762903 EFZ20550.1 KZ288292 PBC29085.1 GL449613 EFN82454.1 GFXV01003710 MBW15515.1 ABLF02039909 GGMR01002966 MBY15585.1 GGMS01001883 MBY71086.1 AJ271674 CAC00652.1 JR053189 AEY61871.1 GBXI01012766 GBXI01007088 JAD01526.1 JAD07204.1 KQ414628 KOC67375.1 GBYB01006111 JAG75878.1 GDHF01025164 GDHF01022334 GDHF01021782 GDHF01020628 GDHF01019942 GDHF01013839 GDHF01009740 GDHF01007804 GDHF01006993 GDHF01006140 GDHF01004604 JAI27150.1 JAI29980.1 JAI30532.1 JAI31686.1 JAI32372.1 JAI38475.1 JAI42574.1 JAI44510.1 JAI45321.1 JAI46174.1 JAI47710.1 KQ434844 KZC08162.1 AAZO01003698 DS235324 EEB14676.1 GFTR01005496 JAW10930.1 GBBI01002143 JAC16569.1 KQ435794 KOX74139.1 NEVH01025127 PNF16136.1 ACPB03017770 GBHO01004044 GBRD01009005 GDHC01004987 JAG39560.1 JAG56816.1 JAQ13642.1 KK852463 KDR23367.1 GEBQ01022333 GEBQ01014002 JAT17644.1 JAT25975.1 GECZ01005582 GECZ01000688 JAS64187.1 JAS69081.1 GAKP01016379 GAKP01016378 JAC42574.1 AXCN02000029 GL732555 EFX78769.1 GALA01001014 JAA93838.1 GDIP01028209 JAM75506.1 KF516617 AII16521.1 GDIQ01085762 JAN08975.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000192223 UP000007266 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000002320 UP000001292 UP000076407 UP000075884 UP000075883 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000092443 UP000000673 UP000075880 UP000078200 UP000008744 UP000092445 UP000092460 UP000192221 UP000008792 UP000009192 UP000001819 UP000002282 UP000007798 UP000001070 UP000008711 UP000000304 UP000007801 UP000053097 UP000279307 UP000000803 UP000078492 UP000007755 UP000078540 UP000036403 UP000078541 UP000005205 UP000075809 UP000095301 UP000095300 UP000242457 UP000008237 UP000007819 UP000005203 UP000053825 UP000076502 UP000009046 UP000053105 UP000235965 UP000015103 UP000027135 UP000079169 UP000075886 UP000000305 UP000085678
UP000192223 UP000007266 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000002320 UP000001292 UP000076407 UP000075884 UP000075883 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000092443 UP000000673 UP000075880 UP000078200 UP000008744 UP000092445 UP000092460 UP000192221 UP000008792 UP000009192 UP000001819 UP000002282 UP000007798 UP000001070 UP000008711 UP000000304 UP000007801 UP000053097 UP000279307 UP000000803 UP000078492 UP000007755 UP000078540 UP000036403 UP000078541 UP000005205 UP000075809 UP000095301 UP000095300 UP000242457 UP000008237 UP000007819 UP000005203 UP000053825 UP000076502 UP000009046 UP000053105 UP000235965 UP000015103 UP000027135 UP000079169 UP000075886 UP000000305 UP000085678
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A5JNM1
A0A0S2VHW6
A0A2A4K291
A0A194RME0
A0A212FLD3
A0A194PMX1
+ More
A0A0L7L866 A0A1W4WWQ6 D6WC07 A0A1Y1MUE8 N6UW64 A0A1L8DM17 A0A0K8TSM1 A0A1L8DKK8 A0A1L8DLK2 T1DEF5 A0A1Q3FP44 Q1HQW4 A0A023EQD8 A0A0C9RJF4 B0XFG0 B4HW89 A0A1Y9IZU8 A0A3F2YVZ5 U5EXF6 A0A182LYM3 A0A182SXB9 A0A182VLF0 A0A182TV87 A0A182L915 Q7PXF7 A0A1A9XGX3 T1E804 A0A2M3ZG62 A0A2M4A7I4 W5JN41 A0A182JM51 A0A1A9UCN4 B4G7Y8 A0A1B0A4X0 A0A1B0C119 A0A1W4W8R0 B4M9H6 B4KH13 Q29NB1 B4NY66 B4MVA0 B4JCE3 B3N8G6 B4Q7V0 B3MUX5 A0A026WR62 P12370 A0A0G4AJS6 W8C0H4 A0A195EJF1 A0A2R7VYK8 F4X2B5 A0A195BCI3 A0A0J7KSG5 A0A195FN55 A0A158NJP0 A0A151X6H1 T1PGE9 A0A1I8NM79 E9IFT9 A0A2A3EBH8 E2BPC9 A0A2H8TN08 J9JK31 A0A2S2NEM0 A0A2S2PZY9 Q9NAS6 V9IMJ8 A0A087ZX62 A0A0A1X7C9 A0A0L7R929 A0A0C9RGE4 A0A0K8V0C2 A0A154P8G8 E0VMS0 A0A224XR64 A0A023F5V8 A0A0M9A1X7 A0A2J7PIG0 T1IA19 A0A0A9ZCM8 A0A067RHU0 A0A1B6LQI5 A0A1B6GP45 A0A1S3DFC7 A0A034VJA2 A0A182QBI4 E9GNI4 A0A1S3JH22 T1E265 A0A1S3JGA5 A0A0P6AK16 A0A076FKS1 A0A0P6DJB0
A0A0L7L866 A0A1W4WWQ6 D6WC07 A0A1Y1MUE8 N6UW64 A0A1L8DM17 A0A0K8TSM1 A0A1L8DKK8 A0A1L8DLK2 T1DEF5 A0A1Q3FP44 Q1HQW4 A0A023EQD8 A0A0C9RJF4 B0XFG0 B4HW89 A0A1Y9IZU8 A0A3F2YVZ5 U5EXF6 A0A182LYM3 A0A182SXB9 A0A182VLF0 A0A182TV87 A0A182L915 Q7PXF7 A0A1A9XGX3 T1E804 A0A2M3ZG62 A0A2M4A7I4 W5JN41 A0A182JM51 A0A1A9UCN4 B4G7Y8 A0A1B0A4X0 A0A1B0C119 A0A1W4W8R0 B4M9H6 B4KH13 Q29NB1 B4NY66 B4MVA0 B4JCE3 B3N8G6 B4Q7V0 B3MUX5 A0A026WR62 P12370 A0A0G4AJS6 W8C0H4 A0A195EJF1 A0A2R7VYK8 F4X2B5 A0A195BCI3 A0A0J7KSG5 A0A195FN55 A0A158NJP0 A0A151X6H1 T1PGE9 A0A1I8NM79 E9IFT9 A0A2A3EBH8 E2BPC9 A0A2H8TN08 J9JK31 A0A2S2NEM0 A0A2S2PZY9 Q9NAS6 V9IMJ8 A0A087ZX62 A0A0A1X7C9 A0A0L7R929 A0A0C9RGE4 A0A0K8V0C2 A0A154P8G8 E0VMS0 A0A224XR64 A0A023F5V8 A0A0M9A1X7 A0A2J7PIG0 T1IA19 A0A0A9ZCM8 A0A067RHU0 A0A1B6LQI5 A0A1B6GP45 A0A1S3DFC7 A0A034VJA2 A0A182QBI4 E9GNI4 A0A1S3JH22 T1E265 A0A1S3JGA5 A0A0P6AK16 A0A076FKS1 A0A0P6DJB0
PDB
3MVJ
E-value=6.67985e-168,
Score=1515
Ontologies
PATHWAY
GO
GO:0004674
GO:0005524
GO:0007314
GO:0071361
GO:2000249
GO:0005829
GO:0040040
GO:0045880
GO:0008103
GO:0010628
GO:0045879
GO:0007317
GO:0030425
GO:0007448
GO:0007622
GO:0008355
GO:0008359
GO:0005886
GO:0048682
GO:0044297
GO:0046823
GO:0007615
GO:0050804
GO:0045187
GO:0007476
GO:0032436
GO:0042981
GO:0048477
GO:0006468
GO:0048749
GO:0019933
GO:0048149
GO:0004691
GO:0004672
GO:0005515
GO:0016787
GO:0004713
GO:0016491
GO:0015930
PANTHER
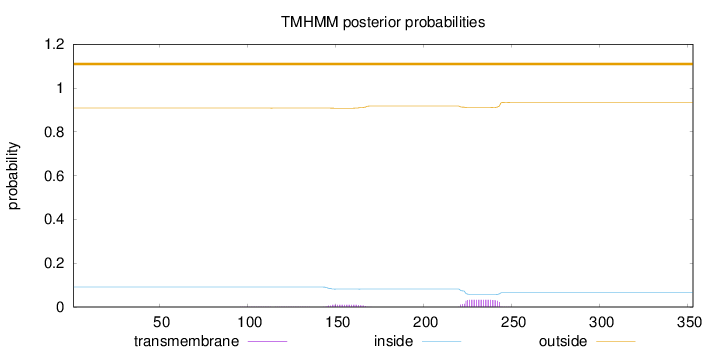
Topology
Length:
353
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.87907
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.09163
outside
1 - 353
Population Genetic Test Statistics
Pi
190.714269
Theta
216.014928
Tajima's D
-0.412565
CLR
0.089181
CSRT
0.261586920653967
Interpretation
Uncertain
