Pre Gene Modal
BGIBMGA011586
Annotation
UPF0414_transmembrane_protein-like_protein_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 2.485
Sequence
CDS
ATGTCCAGGCTACGATTTGGCAGATCGAACGACGGATATCGTCTCGTATCCAATGATGATAGTGGTTTCTCTGACGCTCAGTTTGAAGCACCACCACCGAAGATTCCATGGAAAGCCATAACATTAGCTGCATTTCTGTTTTTGGTCGGGACAATCCTTCTTATAGTTGGAAGCTTGATAGTGACTGGTCACATTGACACAAAATATTCAGATAGGTTGTGGCCTATGATCATTTTAGGAGGCATAATGTTTCTTCCTGGAGCTTATCACATAAGGATAGCGTACTGCGCATATAAGCAGTATCCAGGATATAGTTTTACAGACATTCCAGAATTTGATTAG
Protein
MSRLRFGRSNDGYRLVSNDDSGFSDAQFEAPPPKIPWKAITLAAFLFLVGTILLIVGSLIVTGHIDTKYSDRLWPMIILGGIMFLPGAYHIRIAYCAYKQYPGYSFTDIPEFD
Summary
Uniprot
H9JPY1
A0A3S2TKG0
A0A2H1VVT8
A0A0L7LUZ8
A0A1E1W7Q0
A0A194PKW2
+ More
A0A212FJU9 S4P2K1 E2C2C8 A0A310SET0 A0A0L7RH81 A0A232ET73 A0A088ALX5 A0A2A3E6T5 V9IJ24 A0A0M9A519 E2A1A4 A0A195D484 A0A195FYC9 A0A158NW03 A0A0T6AT70 A0A3L8DRF9 A0A0L8GKZ3 A0A139WN02 D6WBE8 A0A139WNF0 A0A026VZI8 A0A2B4RXE2 A0A3S0ZE22 A7SGM3 V3YXQ9 A0A3M6UN94 A0A210PYE6 A0A2J7QSK5 E9IHP0 A0A2C9JS79 R7TIJ4 A0A067QKY9 A0A293MVV7 E9FX17 A0A1S3H695 E0VYH1 T1HHR5 A0A0P4VG39 A0A0L8GL64 U5EXC0 A0A087UXR2 T1ISH7 A0A1B6I2J4 A0A2T7P7J2 A0A1B6ETI5 A0A1B6GED5 A0A1B6H2C1 B7QFU0 A0A1D2NJ41 A0A023G805 G3MQP7 A0A0N5DPI7 A0A3Q2DAI5 Q68EJ3 A0A3Q3IRE2 A0A131XHK2 A0A085ME37 A0A3B3H4T2 A0A3P9JN44 W4YZB9 T2M8D7 A0A0P5H910 A0A3P8UHB4 A0A077Z0A1 A0A164VF87 A0A0N7ZNE2 A0A0N8AFS2 A0A224YZF5 A0A131Z1C2 A0A3P8WP49 A0A0N8DV25 A0A146ZL98 K4G0B7 A0A3Q3IHX7 C3Z0I7 L7M5U7 A0A0P6I659 V9L867 H2ZK77 B5X879 A0A0S7LRK3 A0A0E9X6P3 A0A3B3Z193 A0A3B3TY01 A0A1W4YFA7 A0A3Q3X078 M3ZIJ8 A0A060VWQ0 V9L8V3 A0A0C9S9K9 H2RR19 A0A087YAH9 C1BJ42 C1BX22
A0A212FJU9 S4P2K1 E2C2C8 A0A310SET0 A0A0L7RH81 A0A232ET73 A0A088ALX5 A0A2A3E6T5 V9IJ24 A0A0M9A519 E2A1A4 A0A195D484 A0A195FYC9 A0A158NW03 A0A0T6AT70 A0A3L8DRF9 A0A0L8GKZ3 A0A139WN02 D6WBE8 A0A139WNF0 A0A026VZI8 A0A2B4RXE2 A0A3S0ZE22 A7SGM3 V3YXQ9 A0A3M6UN94 A0A210PYE6 A0A2J7QSK5 E9IHP0 A0A2C9JS79 R7TIJ4 A0A067QKY9 A0A293MVV7 E9FX17 A0A1S3H695 E0VYH1 T1HHR5 A0A0P4VG39 A0A0L8GL64 U5EXC0 A0A087UXR2 T1ISH7 A0A1B6I2J4 A0A2T7P7J2 A0A1B6ETI5 A0A1B6GED5 A0A1B6H2C1 B7QFU0 A0A1D2NJ41 A0A023G805 G3MQP7 A0A0N5DPI7 A0A3Q2DAI5 Q68EJ3 A0A3Q3IRE2 A0A131XHK2 A0A085ME37 A0A3B3H4T2 A0A3P9JN44 W4YZB9 T2M8D7 A0A0P5H910 A0A3P8UHB4 A0A077Z0A1 A0A164VF87 A0A0N7ZNE2 A0A0N8AFS2 A0A224YZF5 A0A131Z1C2 A0A3P8WP49 A0A0N8DV25 A0A146ZL98 K4G0B7 A0A3Q3IHX7 C3Z0I7 L7M5U7 A0A0P6I659 V9L867 H2ZK77 B5X879 A0A0S7LRK3 A0A0E9X6P3 A0A3B3Z193 A0A3B3TY01 A0A1W4YFA7 A0A3Q3X078 M3ZIJ8 A0A060VWQ0 V9L8V3 A0A0C9S9K9 H2RR19 A0A087YAH9 C1BJ42 C1BX22
Pubmed
19121390
26227816
26354079
22118469
23622113
20798317
+ More
28648823 21347285 30249741 18362917 19820115 24508170 17615350 23254933 30382153 28812685 21282665 15562597 24845553 21292972 20566863 27129103 27289101 22216098 23594743 28049606 24929829 17554307 24065732 24487278 28797301 26830274 23056606 18563158 25576852 24402279 20433749 25613341 23542700 24755649 26131772 21551351 25069045
28648823 21347285 30249741 18362917 19820115 24508170 17615350 23254933 30382153 28812685 21282665 15562597 24845553 21292972 20566863 27129103 27289101 22216098 23594743 28049606 24929829 17554307 24065732 24487278 28797301 26830274 23056606 18563158 25576852 24402279 20433749 25613341 23542700 24755649 26131772 21551351 25069045
EMBL
BABH01015513
RSAL01000081
RVE48562.1
ODYU01004734
SOQ44923.1
JTDY01000071
+ More
KOB79036.1 GDQN01008079 JAT82975.1 KQ459601 KPI94071.1 AGBW02008229 OWR53980.1 GAIX01009967 JAA82593.1 GL452124 EFN77878.1 KQ760561 OAD59791.1 KQ414590 KOC70332.1 NNAY01002322 OXU21551.1 KZ288370 PBC26751.1 JR048364 AEY60642.1 KQ435753 KOX76093.1 GL435730 EFN72798.1 KQ976870 KYN07697.1 KQ981169 KYN45312.1 ADTU01027611 LJIG01022876 KRT78276.1 QOIP01000005 RLU23025.1 KQ421356 KOF77661.1 KQ971312 KYB29398.1 EEZ98724.1 KYB29397.1 KK107538 EZA49152.1 LSMT01000275 PFX21469.1 RQTK01000635 RUS76792.1 DS469654 EDO37093.1 KB203854 ESO82838.1 RCHS01001119 RMX55136.1 NEDP02005386 OWF41502.1 NEVH01011876 PNF31566.1 GL763289 EFZ19906.1 AMQN01002608 KB309694 ELT93559.1 KK853313 KDR08632.1 GFWV01018722 MAA43450.1 GL732526 EFX88337.1 DS235845 EEB18427.1 ACPB03019965 GDKW01003501 JAI53094.1 KOF77663.1 GANO01002608 JAB57263.1 KK122182 KFM82151.1 JH431432 GECU01026549 GECU01010011 JAS81157.1 JAS97695.1 PZQS01000005 PVD29395.1 GECZ01028568 GECZ01022808 GECZ01018191 GECZ01014570 GECZ01013313 GECZ01006475 JAS41201.1 JAS46961.1 JAS51578.1 JAS55199.1 JAS56456.1 JAS63294.1 GECZ01008970 JAS60799.1 GECZ01028901 GECZ01000940 JAS40868.1 JAS68829.1 ABJB010832055 DS927877 EEC17712.1 LJIJ01000026 ODN05271.1 GBBM01006388 JAC29030.1 JO844198 AEO35815.1 AL928672 BC080236 AAH80236.1 GEFH01002983 JAP65598.1 KL363199 KFD55483.1 AAGJ04077390 HAAD01002005 CDG68237.1 GDIQ01229769 JAK21956.1 HG805854 CDW53471.1 LRGB01001361 KZS12264.1 GDIP01228401 JAI95000.1 GDIP01151761 JAJ71641.1 GFPF01008557 MAA19703.1 GEDV01004007 JAP84550.1 GDIQ01089068 JAN05669.1 GCES01019618 JAR66705.1 JX052758 AFK10986.1 GG666568 EEN54035.1 GACK01006511 JAA58523.1 GDIQ01016232 JAN78505.1 JW875473 AFP07990.1 BT047248 BT125152 ACI67049.1 ADM15893.1 GBYX01153449 JAO92063.1 GBXM01010318 JAH98259.1 FR904268 CDQ56740.1 JW875900 AFP08417.1 GBZX01003128 JAG89612.1 AYCK01000971 BT074621 BT074689 ACO09045.1 BT079151 ACO13575.1
KOB79036.1 GDQN01008079 JAT82975.1 KQ459601 KPI94071.1 AGBW02008229 OWR53980.1 GAIX01009967 JAA82593.1 GL452124 EFN77878.1 KQ760561 OAD59791.1 KQ414590 KOC70332.1 NNAY01002322 OXU21551.1 KZ288370 PBC26751.1 JR048364 AEY60642.1 KQ435753 KOX76093.1 GL435730 EFN72798.1 KQ976870 KYN07697.1 KQ981169 KYN45312.1 ADTU01027611 LJIG01022876 KRT78276.1 QOIP01000005 RLU23025.1 KQ421356 KOF77661.1 KQ971312 KYB29398.1 EEZ98724.1 KYB29397.1 KK107538 EZA49152.1 LSMT01000275 PFX21469.1 RQTK01000635 RUS76792.1 DS469654 EDO37093.1 KB203854 ESO82838.1 RCHS01001119 RMX55136.1 NEDP02005386 OWF41502.1 NEVH01011876 PNF31566.1 GL763289 EFZ19906.1 AMQN01002608 KB309694 ELT93559.1 KK853313 KDR08632.1 GFWV01018722 MAA43450.1 GL732526 EFX88337.1 DS235845 EEB18427.1 ACPB03019965 GDKW01003501 JAI53094.1 KOF77663.1 GANO01002608 JAB57263.1 KK122182 KFM82151.1 JH431432 GECU01026549 GECU01010011 JAS81157.1 JAS97695.1 PZQS01000005 PVD29395.1 GECZ01028568 GECZ01022808 GECZ01018191 GECZ01014570 GECZ01013313 GECZ01006475 JAS41201.1 JAS46961.1 JAS51578.1 JAS55199.1 JAS56456.1 JAS63294.1 GECZ01008970 JAS60799.1 GECZ01028901 GECZ01000940 JAS40868.1 JAS68829.1 ABJB010832055 DS927877 EEC17712.1 LJIJ01000026 ODN05271.1 GBBM01006388 JAC29030.1 JO844198 AEO35815.1 AL928672 BC080236 AAH80236.1 GEFH01002983 JAP65598.1 KL363199 KFD55483.1 AAGJ04077390 HAAD01002005 CDG68237.1 GDIQ01229769 JAK21956.1 HG805854 CDW53471.1 LRGB01001361 KZS12264.1 GDIP01228401 JAI95000.1 GDIP01151761 JAJ71641.1 GFPF01008557 MAA19703.1 GEDV01004007 JAP84550.1 GDIQ01089068 JAN05669.1 GCES01019618 JAR66705.1 JX052758 AFK10986.1 GG666568 EEN54035.1 GACK01006511 JAA58523.1 GDIQ01016232 JAN78505.1 JW875473 AFP07990.1 BT047248 BT125152 ACI67049.1 ADM15893.1 GBYX01153449 JAO92063.1 GBXM01010318 JAH98259.1 FR904268 CDQ56740.1 JW875900 AFP08417.1 GBZX01003128 JAG89612.1 AYCK01000971 BT074621 BT074689 ACO09045.1 BT079151 ACO13575.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000053268
UP000007151
UP000008237
+ More
UP000053825 UP000215335 UP000005203 UP000242457 UP000053105 UP000000311 UP000078542 UP000078541 UP000005205 UP000279307 UP000053454 UP000007266 UP000053097 UP000225706 UP000271974 UP000001593 UP000030746 UP000275408 UP000242188 UP000235965 UP000076420 UP000014760 UP000027135 UP000000305 UP000085678 UP000009046 UP000015103 UP000054359 UP000245119 UP000001555 UP000094527 UP000046395 UP000265020 UP000000437 UP000261600 UP000001038 UP000265180 UP000265200 UP000007110 UP000265120 UP000030665 UP000076858 UP000001554 UP000007875 UP000087266 UP000261480 UP000261500 UP000192224 UP000261620 UP000002852 UP000193380 UP000005226 UP000028760 UP000265140
UP000053825 UP000215335 UP000005203 UP000242457 UP000053105 UP000000311 UP000078542 UP000078541 UP000005205 UP000279307 UP000053454 UP000007266 UP000053097 UP000225706 UP000271974 UP000001593 UP000030746 UP000275408 UP000242188 UP000235965 UP000076420 UP000014760 UP000027135 UP000000305 UP000085678 UP000009046 UP000015103 UP000054359 UP000245119 UP000001555 UP000094527 UP000046395 UP000265020 UP000000437 UP000261600 UP000001038 UP000265180 UP000265200 UP000007110 UP000265120 UP000030665 UP000076858 UP000001554 UP000007875 UP000087266 UP000261480 UP000261500 UP000192224 UP000261620 UP000002852 UP000193380 UP000005226 UP000028760 UP000265140
PRIDE
Pfam
PF05915 DUF872
Interpro
IPR008590
DUF872_TM
ProteinModelPortal
H9JPY1
A0A3S2TKG0
A0A2H1VVT8
A0A0L7LUZ8
A0A1E1W7Q0
A0A194PKW2
+ More
A0A212FJU9 S4P2K1 E2C2C8 A0A310SET0 A0A0L7RH81 A0A232ET73 A0A088ALX5 A0A2A3E6T5 V9IJ24 A0A0M9A519 E2A1A4 A0A195D484 A0A195FYC9 A0A158NW03 A0A0T6AT70 A0A3L8DRF9 A0A0L8GKZ3 A0A139WN02 D6WBE8 A0A139WNF0 A0A026VZI8 A0A2B4RXE2 A0A3S0ZE22 A7SGM3 V3YXQ9 A0A3M6UN94 A0A210PYE6 A0A2J7QSK5 E9IHP0 A0A2C9JS79 R7TIJ4 A0A067QKY9 A0A293MVV7 E9FX17 A0A1S3H695 E0VYH1 T1HHR5 A0A0P4VG39 A0A0L8GL64 U5EXC0 A0A087UXR2 T1ISH7 A0A1B6I2J4 A0A2T7P7J2 A0A1B6ETI5 A0A1B6GED5 A0A1B6H2C1 B7QFU0 A0A1D2NJ41 A0A023G805 G3MQP7 A0A0N5DPI7 A0A3Q2DAI5 Q68EJ3 A0A3Q3IRE2 A0A131XHK2 A0A085ME37 A0A3B3H4T2 A0A3P9JN44 W4YZB9 T2M8D7 A0A0P5H910 A0A3P8UHB4 A0A077Z0A1 A0A164VF87 A0A0N7ZNE2 A0A0N8AFS2 A0A224YZF5 A0A131Z1C2 A0A3P8WP49 A0A0N8DV25 A0A146ZL98 K4G0B7 A0A3Q3IHX7 C3Z0I7 L7M5U7 A0A0P6I659 V9L867 H2ZK77 B5X879 A0A0S7LRK3 A0A0E9X6P3 A0A3B3Z193 A0A3B3TY01 A0A1W4YFA7 A0A3Q3X078 M3ZIJ8 A0A060VWQ0 V9L8V3 A0A0C9S9K9 H2RR19 A0A087YAH9 C1BJ42 C1BX22
A0A212FJU9 S4P2K1 E2C2C8 A0A310SET0 A0A0L7RH81 A0A232ET73 A0A088ALX5 A0A2A3E6T5 V9IJ24 A0A0M9A519 E2A1A4 A0A195D484 A0A195FYC9 A0A158NW03 A0A0T6AT70 A0A3L8DRF9 A0A0L8GKZ3 A0A139WN02 D6WBE8 A0A139WNF0 A0A026VZI8 A0A2B4RXE2 A0A3S0ZE22 A7SGM3 V3YXQ9 A0A3M6UN94 A0A210PYE6 A0A2J7QSK5 E9IHP0 A0A2C9JS79 R7TIJ4 A0A067QKY9 A0A293MVV7 E9FX17 A0A1S3H695 E0VYH1 T1HHR5 A0A0P4VG39 A0A0L8GL64 U5EXC0 A0A087UXR2 T1ISH7 A0A1B6I2J4 A0A2T7P7J2 A0A1B6ETI5 A0A1B6GED5 A0A1B6H2C1 B7QFU0 A0A1D2NJ41 A0A023G805 G3MQP7 A0A0N5DPI7 A0A3Q2DAI5 Q68EJ3 A0A3Q3IRE2 A0A131XHK2 A0A085ME37 A0A3B3H4T2 A0A3P9JN44 W4YZB9 T2M8D7 A0A0P5H910 A0A3P8UHB4 A0A077Z0A1 A0A164VF87 A0A0N7ZNE2 A0A0N8AFS2 A0A224YZF5 A0A131Z1C2 A0A3P8WP49 A0A0N8DV25 A0A146ZL98 K4G0B7 A0A3Q3IHX7 C3Z0I7 L7M5U7 A0A0P6I659 V9L867 H2ZK77 B5X879 A0A0S7LRK3 A0A0E9X6P3 A0A3B3Z193 A0A3B3TY01 A0A1W4YFA7 A0A3Q3X078 M3ZIJ8 A0A060VWQ0 V9L8V3 A0A0C9S9K9 H2RR19 A0A087YAH9 C1BJ42 C1BX22
Ontologies
GO
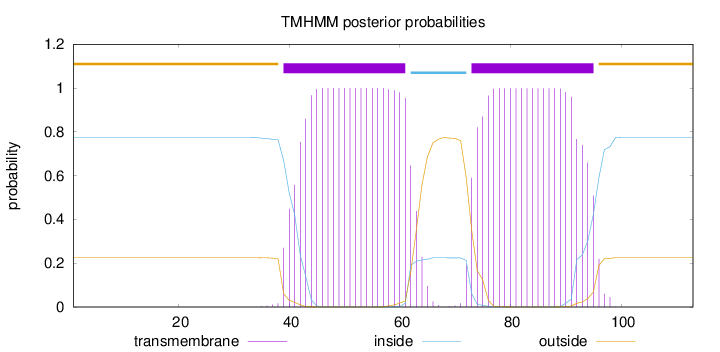
Topology
Length:
113
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.63791
Exp number, first 60 AAs:
19.85245
Total prob of N-in:
0.77495
POSSIBLE N-term signal
sequence
outside
1 - 38
TMhelix
39 - 61
inside
62 - 72
TMhelix
73 - 95
outside
96 - 113
Population Genetic Test Statistics
Pi
200.952434
Theta
197.939589
Tajima's D
0.06324
CLR
0
CSRT
0.387530623468827
Interpretation
Uncertain
