Gene
KWMTBOMO13886
Pre Gene Modal
BGIBMGA011436
Annotation
PREDICTED:_uncharacterized_protein_LOC106708730_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.961
Sequence
CDS
ATGGAAATCGTGCCAGAAGGGAAAAACTTTCGAATAGTTCCTGAAGACGAACTTCAAGCGGTGGCCGATTTCTTAGTGCGGTACATGCCTGAATCTCTAAAGTTTCATCAGACGATTCAAACATATCTAAACAACAAAGTCTGGAACTTCCATTTTTATGTAGCGAAGAATTGGCCAGAGGACCCGATTTGCCTACATTTTCCCGGATGTACGAGCACGCCAAACAGTCAAATTTACGAAAGCGTGACGGTGTTTTGTCCATCCGCCCGGGCTGATCTAGTTGATCTGGTGACTTCAGAAAGCATCCTACTAGATTTAACTCAACCATTGTACCTAAATTTCACTCACGAAGCTATCGTTAATCGCTTCGAGAAGCGCTATGAGGCCCACGATAAGATCACCGGCGATGTTTATGCCTGTGACAATCCACCGGCGGATGTCGATACTGATGGATTGCCACCGGACGTGGAGTTAGTGCGCTTGCAGCCGGAGCACATGAAGGCCGTGCACGATCTTTATCCGGCAAGTGATATCGAATGTCGTGAGGTATTTGAGAAGTTGGTAGCCGAACTACCGGCCTACGGGATCTTTGTCGAAGGTCAACTTGCCGCTTGGATGGTTCAATCATACTACGGAGCCATGTTCTCTATGCAAACTCGCCCCGAATTCCGTAGGAAAGGTTACGGTATTTACTTAGCTCGCCGCCTAACTAAAGAAGTCGCAGCCCGGGGCTATAAGCCCTTCGTCGTCATTCGTCCGGAAAACGACGCGTCCCGTTCGCTTTACACGAAACTCGGCTTTGAGAAACGTTTCCGAACGGTGCGCGCCGTTTTGCGTCCTCGCTAG
Protein
MEIVPEGKNFRIVPEDELQAVADFLVRYMPESLKFHQTIQTYLNNKVWNFHFYVAKNWPEDPICLHFPGCTSTPNSQIYESVTVFCPSARADLVDLVTSESILLDLTQPLYLNFTHEAIVNRFEKRYEAHDKITGDVYACDNPPADVDTDGLPPDVELVRLQPEHMKAVHDLYPASDIECREVFEKLVAELPAYGIFVEGQLAAWMVQSYYGAMFSMQTRPEFRRKGYGIYLARRLTKEVAARGYKPFVVIRPENDASRSLYTKLGFEKRFRTVRAVLRPR
Summary
Uniprot
A0A2A4JIV9
A0A194PMW1
A0A3S2NZK4
A0A212FAY6
A0A194RMD1
H9JPI1
+ More
A0A0L7LK05 A0A2H1WUA2 A0A1B0CIK4 A0A1L8DBI1 A0A1B6DVR1 A0A1B6MQD7 A0A1Y1NG75 A0A0A1WK00 A0A0A1XS72 A0A034VRW7 A0A0K8V399 W8AXP4 B3N4A0 A0A067R7Z4 A0A0L0C921 Q9VR30 A0A0J9QVP2 A0A1W4UIB3 B4I3C8 D6WBE7 B4NZ12 B4Q330 A0A1A9ZR26 B3ML93 B4G8X4 A0A1B0G793 B5DIA5 A0A0R3NZP7 A0A1I8P1P6 A0A1J1J079 A0A0M4END9 B4N0H6 A0A3B0JF25 B4LTU6 B4KGC3 A0A0Q9WKK6 U4UV73 A0A1W4XJD1 A0A1I8MN05 B4JAR4 E2C2C9 A0A0N0U5U1 A0A3L8DRQ4 A0A026VZE3 K7J9P5 A0A3Q0JMQ3 Q16US5 A0A1S4FNM2 A0A182QVH6 A0A1Q3G521 V9IJH4 A0A2A3E4Y2 A0A182IVQ0 A0A182YN28 A0A182RAG9 A0A182K9Y4 A0A182VSQ6 A0A0C9RGJ7 A0A182XPF5 E2A1A3 A0A158NW04 Q7PTY3 A0A232ETN8 A0A195FXC1 A0A182KLQ5 A0A182FQI0 A0A195D483 A0A1A9XUE5 W5JBH8 A0A1A9UGU4 A0A154P950 A0A1B0DBU7 A0A2A3E6F8 E0VJ50 A0A0L7RHV4 A0A088ALY8 F4WN28 A0A195BN18 A0A2R7X0J3 A0A2J7QCM8 A0A1B6IF95 J3JU19 A0A1B6DTN2 A0A310SNS0 A0A2P8YX19 A0A0J7NL86 A0A336LWW3 A0A182N8Y5 A0A1W4X951 A0A084W5N5 A0A182PWL3 A0A151JN63
A0A0L7LK05 A0A2H1WUA2 A0A1B0CIK4 A0A1L8DBI1 A0A1B6DVR1 A0A1B6MQD7 A0A1Y1NG75 A0A0A1WK00 A0A0A1XS72 A0A034VRW7 A0A0K8V399 W8AXP4 B3N4A0 A0A067R7Z4 A0A0L0C921 Q9VR30 A0A0J9QVP2 A0A1W4UIB3 B4I3C8 D6WBE7 B4NZ12 B4Q330 A0A1A9ZR26 B3ML93 B4G8X4 A0A1B0G793 B5DIA5 A0A0R3NZP7 A0A1I8P1P6 A0A1J1J079 A0A0M4END9 B4N0H6 A0A3B0JF25 B4LTU6 B4KGC3 A0A0Q9WKK6 U4UV73 A0A1W4XJD1 A0A1I8MN05 B4JAR4 E2C2C9 A0A0N0U5U1 A0A3L8DRQ4 A0A026VZE3 K7J9P5 A0A3Q0JMQ3 Q16US5 A0A1S4FNM2 A0A182QVH6 A0A1Q3G521 V9IJH4 A0A2A3E4Y2 A0A182IVQ0 A0A182YN28 A0A182RAG9 A0A182K9Y4 A0A182VSQ6 A0A0C9RGJ7 A0A182XPF5 E2A1A3 A0A158NW04 Q7PTY3 A0A232ETN8 A0A195FXC1 A0A182KLQ5 A0A182FQI0 A0A195D483 A0A1A9XUE5 W5JBH8 A0A1A9UGU4 A0A154P950 A0A1B0DBU7 A0A2A3E6F8 E0VJ50 A0A0L7RHV4 A0A088ALY8 F4WN28 A0A195BN18 A0A2R7X0J3 A0A2J7QCM8 A0A1B6IF95 J3JU19 A0A1B6DTN2 A0A310SNS0 A0A2P8YX19 A0A0J7NL86 A0A336LWW3 A0A182N8Y5 A0A1W4X951 A0A084W5N5 A0A182PWL3 A0A151JN63
Pubmed
26354079
22118469
19121390
26227816
28004739
25830018
+ More
25348373 24495485 17994087 24845553 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 17550304 15632085 23537049 25315136 20798317 30249741 24508170 20075255 17510324 25244985 21347285 9087549 12364791 14747013 17210077 28648823 20966253 20920257 23761445 20566863 21719571 22516182 29403074 24438588
25348373 24495485 17994087 24845553 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 17550304 15632085 23537049 25315136 20798317 30249741 24508170 20075255 17510324 25244985 21347285 9087549 12364791 14747013 17210077 28648823 20966253 20920257 23761445 20566863 21719571 22516182 29403074 24438588
EMBL
NWSH01001408
PCG71350.1
KQ459601
KPI94069.1
RSAL01000081
RVE48560.1
+ More
AGBW02009400 OWR50905.1 KQ460205 KPJ17146.1 BABH01015508 JTDY01000782 KOB75898.1 ODYU01011110 SOQ56641.1 AJWK01013486 AJWK01013487 AJWK01013488 GFDF01010400 JAV03684.1 GEDC01007545 JAS29753.1 GEBQ01001868 JAT38109.1 GEZM01003823 GEZM01003821 JAV96538.1 GBXI01015110 GBXI01007229 GBXI01000696 JAC99181.1 JAD07063.1 JAD13596.1 GBXI01000446 JAD13846.1 GAKP01012896 JAC46056.1 GDHF01019244 JAI33070.1 GAMC01012807 GAMC01012805 JAB93750.1 CH954177 EDV57770.1 KK852643 KDR19561.1 JRES01000753 KNC28742.1 AE014134 BT001689 AAF50978.3 AAN71444.1 CM002910 KMY88122.1 CH480820 EDW54273.1 KQ971312 EEZ97892.1 CM000157 EDW87676.1 CM000361 EDX03746.1 CH902620 EDV31711.2 CH479180 EDW28804.1 CCAG010022296 CH379060 EDY70045.2 KRT04070.1 CVRI01000063 CRL04241.1 CP012523 ALC38175.1 CH963920 EDW77589.1 OUUW01000004 SPP79273.1 CH940649 EDW63997.1 CH933807 EDW11110.2 KRF81387.1 KB632379 ERL94095.1 CH916368 EDW03872.1 GL452124 EFN77879.1 KQ435753 KOX76094.1 QOIP01000005 RLU22478.1 KK107538 EZA49153.1 AAZX01008083 AAZX01012135 AAZX01017200 CH477615 EAT38262.1 AXCN02001877 GFDL01000144 JAV34901.1 JR048940 AEY60807.1 KZ288370 PBC26750.1 GBYB01007490 JAG77257.1 GL435730 EFN72797.1 ADTU01027615 ADTU01027616 AAAB01007675 EAA03049.5 NNAY01002250 OXU21725.1 KQ981169 KYN45315.1 KQ976870 KYN07698.1 ADMH02001972 ETN60119.1 KQ434829 KZC07690.1 AJVK01013796 AJVK01013797 AJVK01013798 PBC26749.1 DS235219 EEB13406.1 KQ414590 KOC70331.1 GL888233 EGI64396.1 KQ976433 KYM87375.1 KK856247 PTY25292.1 NEVH01016289 PNF26334.1 GECU01022133 JAS85573.1 APGK01040393 APGK01040394 APGK01040395 BT126730 KB740979 AEE61692.1 ENN76418.1 GEDC01008261 JAS29037.1 KQ760561 OAD59790.1 PYGN01000309 PSN48797.1 LBMM01003693 KMQ93230.1 UFQT01000262 SSX22546.1 ATLV01020676 KE525304 KFB45529.1 KQ978891 KYN27636.1
AGBW02009400 OWR50905.1 KQ460205 KPJ17146.1 BABH01015508 JTDY01000782 KOB75898.1 ODYU01011110 SOQ56641.1 AJWK01013486 AJWK01013487 AJWK01013488 GFDF01010400 JAV03684.1 GEDC01007545 JAS29753.1 GEBQ01001868 JAT38109.1 GEZM01003823 GEZM01003821 JAV96538.1 GBXI01015110 GBXI01007229 GBXI01000696 JAC99181.1 JAD07063.1 JAD13596.1 GBXI01000446 JAD13846.1 GAKP01012896 JAC46056.1 GDHF01019244 JAI33070.1 GAMC01012807 GAMC01012805 JAB93750.1 CH954177 EDV57770.1 KK852643 KDR19561.1 JRES01000753 KNC28742.1 AE014134 BT001689 AAF50978.3 AAN71444.1 CM002910 KMY88122.1 CH480820 EDW54273.1 KQ971312 EEZ97892.1 CM000157 EDW87676.1 CM000361 EDX03746.1 CH902620 EDV31711.2 CH479180 EDW28804.1 CCAG010022296 CH379060 EDY70045.2 KRT04070.1 CVRI01000063 CRL04241.1 CP012523 ALC38175.1 CH963920 EDW77589.1 OUUW01000004 SPP79273.1 CH940649 EDW63997.1 CH933807 EDW11110.2 KRF81387.1 KB632379 ERL94095.1 CH916368 EDW03872.1 GL452124 EFN77879.1 KQ435753 KOX76094.1 QOIP01000005 RLU22478.1 KK107538 EZA49153.1 AAZX01008083 AAZX01012135 AAZX01017200 CH477615 EAT38262.1 AXCN02001877 GFDL01000144 JAV34901.1 JR048940 AEY60807.1 KZ288370 PBC26750.1 GBYB01007490 JAG77257.1 GL435730 EFN72797.1 ADTU01027615 ADTU01027616 AAAB01007675 EAA03049.5 NNAY01002250 OXU21725.1 KQ981169 KYN45315.1 KQ976870 KYN07698.1 ADMH02001972 ETN60119.1 KQ434829 KZC07690.1 AJVK01013796 AJVK01013797 AJVK01013798 PBC26749.1 DS235219 EEB13406.1 KQ414590 KOC70331.1 GL888233 EGI64396.1 KQ976433 KYM87375.1 KK856247 PTY25292.1 NEVH01016289 PNF26334.1 GECU01022133 JAS85573.1 APGK01040393 APGK01040394 APGK01040395 BT126730 KB740979 AEE61692.1 ENN76418.1 GEDC01008261 JAS29037.1 KQ760561 OAD59790.1 PYGN01000309 PSN48797.1 LBMM01003693 KMQ93230.1 UFQT01000262 SSX22546.1 ATLV01020676 KE525304 KFB45529.1 KQ978891 KYN27636.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000005204
+ More
UP000037510 UP000092461 UP000008711 UP000027135 UP000037069 UP000000803 UP000192221 UP000001292 UP000007266 UP000002282 UP000000304 UP000092445 UP000007801 UP000008744 UP000092444 UP000001819 UP000095300 UP000183832 UP000092553 UP000007798 UP000268350 UP000008792 UP000009192 UP000030742 UP000192223 UP000095301 UP000001070 UP000008237 UP000053105 UP000279307 UP000053097 UP000002358 UP000079169 UP000008820 UP000075886 UP000242457 UP000075880 UP000076408 UP000075900 UP000075881 UP000075920 UP000076407 UP000000311 UP000005205 UP000007062 UP000215335 UP000078541 UP000075882 UP000069272 UP000078542 UP000092443 UP000000673 UP000078200 UP000076502 UP000092462 UP000009046 UP000053825 UP000005203 UP000007755 UP000078540 UP000235965 UP000019118 UP000245037 UP000036403 UP000075884 UP000030765 UP000075885 UP000078492
UP000037510 UP000092461 UP000008711 UP000027135 UP000037069 UP000000803 UP000192221 UP000001292 UP000007266 UP000002282 UP000000304 UP000092445 UP000007801 UP000008744 UP000092444 UP000001819 UP000095300 UP000183832 UP000092553 UP000007798 UP000268350 UP000008792 UP000009192 UP000030742 UP000192223 UP000095301 UP000001070 UP000008237 UP000053105 UP000279307 UP000053097 UP000002358 UP000079169 UP000008820 UP000075886 UP000242457 UP000075880 UP000076408 UP000075900 UP000075881 UP000075920 UP000076407 UP000000311 UP000005205 UP000007062 UP000215335 UP000078541 UP000075882 UP000069272 UP000078542 UP000092443 UP000000673 UP000078200 UP000076502 UP000092462 UP000009046 UP000053825 UP000005203 UP000007755 UP000078540 UP000235965 UP000019118 UP000245037 UP000036403 UP000075884 UP000030765 UP000075885 UP000078492
Pfam
PF08445 FR47
Interpro
SUPFAM
SSF55729
SSF55729
Gene 3D
ProteinModelPortal
A0A2A4JIV9
A0A194PMW1
A0A3S2NZK4
A0A212FAY6
A0A194RMD1
H9JPI1
+ More
A0A0L7LK05 A0A2H1WUA2 A0A1B0CIK4 A0A1L8DBI1 A0A1B6DVR1 A0A1B6MQD7 A0A1Y1NG75 A0A0A1WK00 A0A0A1XS72 A0A034VRW7 A0A0K8V399 W8AXP4 B3N4A0 A0A067R7Z4 A0A0L0C921 Q9VR30 A0A0J9QVP2 A0A1W4UIB3 B4I3C8 D6WBE7 B4NZ12 B4Q330 A0A1A9ZR26 B3ML93 B4G8X4 A0A1B0G793 B5DIA5 A0A0R3NZP7 A0A1I8P1P6 A0A1J1J079 A0A0M4END9 B4N0H6 A0A3B0JF25 B4LTU6 B4KGC3 A0A0Q9WKK6 U4UV73 A0A1W4XJD1 A0A1I8MN05 B4JAR4 E2C2C9 A0A0N0U5U1 A0A3L8DRQ4 A0A026VZE3 K7J9P5 A0A3Q0JMQ3 Q16US5 A0A1S4FNM2 A0A182QVH6 A0A1Q3G521 V9IJH4 A0A2A3E4Y2 A0A182IVQ0 A0A182YN28 A0A182RAG9 A0A182K9Y4 A0A182VSQ6 A0A0C9RGJ7 A0A182XPF5 E2A1A3 A0A158NW04 Q7PTY3 A0A232ETN8 A0A195FXC1 A0A182KLQ5 A0A182FQI0 A0A195D483 A0A1A9XUE5 W5JBH8 A0A1A9UGU4 A0A154P950 A0A1B0DBU7 A0A2A3E6F8 E0VJ50 A0A0L7RHV4 A0A088ALY8 F4WN28 A0A195BN18 A0A2R7X0J3 A0A2J7QCM8 A0A1B6IF95 J3JU19 A0A1B6DTN2 A0A310SNS0 A0A2P8YX19 A0A0J7NL86 A0A336LWW3 A0A182N8Y5 A0A1W4X951 A0A084W5N5 A0A182PWL3 A0A151JN63
A0A0L7LK05 A0A2H1WUA2 A0A1B0CIK4 A0A1L8DBI1 A0A1B6DVR1 A0A1B6MQD7 A0A1Y1NG75 A0A0A1WK00 A0A0A1XS72 A0A034VRW7 A0A0K8V399 W8AXP4 B3N4A0 A0A067R7Z4 A0A0L0C921 Q9VR30 A0A0J9QVP2 A0A1W4UIB3 B4I3C8 D6WBE7 B4NZ12 B4Q330 A0A1A9ZR26 B3ML93 B4G8X4 A0A1B0G793 B5DIA5 A0A0R3NZP7 A0A1I8P1P6 A0A1J1J079 A0A0M4END9 B4N0H6 A0A3B0JF25 B4LTU6 B4KGC3 A0A0Q9WKK6 U4UV73 A0A1W4XJD1 A0A1I8MN05 B4JAR4 E2C2C9 A0A0N0U5U1 A0A3L8DRQ4 A0A026VZE3 K7J9P5 A0A3Q0JMQ3 Q16US5 A0A1S4FNM2 A0A182QVH6 A0A1Q3G521 V9IJH4 A0A2A3E4Y2 A0A182IVQ0 A0A182YN28 A0A182RAG9 A0A182K9Y4 A0A182VSQ6 A0A0C9RGJ7 A0A182XPF5 E2A1A3 A0A158NW04 Q7PTY3 A0A232ETN8 A0A195FXC1 A0A182KLQ5 A0A182FQI0 A0A195D483 A0A1A9XUE5 W5JBH8 A0A1A9UGU4 A0A154P950 A0A1B0DBU7 A0A2A3E6F8 E0VJ50 A0A0L7RHV4 A0A088ALY8 F4WN28 A0A195BN18 A0A2R7X0J3 A0A2J7QCM8 A0A1B6IF95 J3JU19 A0A1B6DTN2 A0A310SNS0 A0A2P8YX19 A0A0J7NL86 A0A336LWW3 A0A182N8Y5 A0A1W4X951 A0A084W5N5 A0A182PWL3 A0A151JN63
PDB
1SQH
E-value=2.75335e-06,
Score=120
Ontologies
GO
Topology
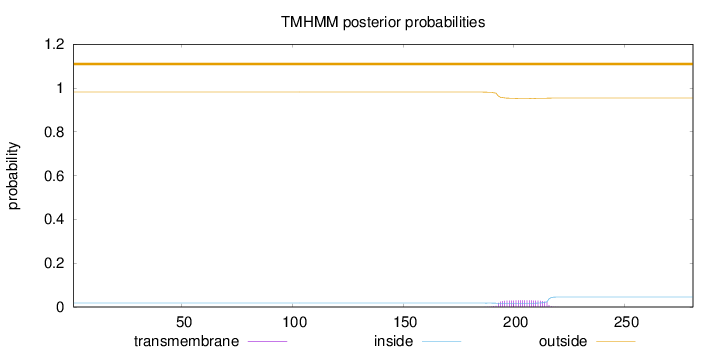
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.679
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01835
outside
1 - 281
Population Genetic Test Statistics
Pi
193.76157
Theta
167.686848
Tajima's D
0.227393
CLR
3.349997
CSRT
0.433978301084946
Interpretation
Uncertain
