Gene
KWMTBOMO13872
Pre Gene Modal
BGIBMGA011579
Annotation
gustatory_receptor_1?_partial_[Helicoverpa_assulta]
Full name
Gustatory receptor
+ More
Gustatory and odorant receptor 21a
Gustatory and odorant receptor 22
Gustatory and odorant receptor 21a
Gustatory and odorant receptor 22
Location in the cell
PlasmaMembrane Reliability : 3.161
Sequence
CDS
ATGCAACCGCAATTTAATCACCGCATTAAGCCGTTTGACGTTTTGTCAGGCCAGTACATTAACTGCAACGCGTTTGGCACGGCGTCCCGGGGCTTGGCAATAAATCTGCACAAGGCTCTAGAGGCGGAACACCCAGCGCTGAAGTTGGCTCAATACCGCCACCTGTGGGTTGATCTGTCGCATATGATGCAGCAATTGGGCCGCGCTTATTCCAACATGTACGGCATTTACTGCATGGTAATATTCTTCACAACAACAATATCATTGTATGGCGCCTTATCGGAAATCTTAGAGCACGGCCTAAGTTACAAGGAAATGGGCTTGTTTGTCATTGTCGCTTACTGCATGACGTTACTCTTCATAATATGTAATGAGGCCTATCACGCTTCGAGAAAGTGTTATAACGGGTAA
Protein
MQPQFNHRIKPFDVLSGQYINCNAFGTASRGLAINLHKALEAEHPALKLAQYRHLWVDLSHMMQQLGRAYSNMYGIYCMVIFFTTTISLYGALSEILEHGLSYKEMGLFVIVAYCMTLLFIICNEAYHASRKCYNG
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Gustatory and odorant receptor which mediates acceptance or avoidance behavior, depending on its substrates. Gr21a and Gr63a together are sufficient for carbon dioxide detection and avoidance behavior. It is possible that the CO(2) receptors Gr63a and Gr21a activate the TRPC channels through Galpha49B and Plc21C. This innate olfactory avoidance behavior can be inhibited by inhibitory interactions of the odors such as 1-hexanol and 2,3-butanedione with Gr21a and Gr63a.
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates. GPRgr22 and GPRgr24 together are sufficient for olfactory carbon dioxide-chemosensation.
Gustatory and odorant receptor which mediates acceptance or avoidance behavior, depending on its substrates. Gr21a and Gr63a together are sufficient for carbon dioxide detection and avoidance behavior. It is possible that the CO(2) receptors Gr63a and Gr21a activate the TRPC channels through Galpha49B and Plc21C. This innate olfactory avoidance behavior can be inhibited by inhibitory interactions of the odors such as 1-hexanol and 2,3-butanedione with Gr21a and Gr63a.
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates. GPRgr22 and GPRgr24 together are sufficient for olfactory carbon dioxide-chemosensation.
Subunit
Gr21a and Gr63a probably form a heterodimer that responds to CO(2).
Similarity
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family. Gr21a subfamily.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family. Gr21a subfamily.
Keywords
Behavior
Cell membrane
Complete proteome
Membrane
Olfaction
Receptor
Reference proteome
Sensory transduction
Transducer
Transmembrane
Transmembrane helix
Glycoprotein
Feature
chain Gustatory and odorant receptor 21a
Uniprot
A0A0B5A3E3
A0A0S1TPL0
H9JPX4
A0A0E4B436
A0A075T806
A0A2A4J8U7
+ More
A0A223HDH7 A0A0V0J1B6 A0A2K8GKX3 A0A0K8TUH7 A0A212ERI7 A0A2H1VDE4 A0A2K8GL76 A0A194RI74 A0A223HD90 A0A194PL61 A0A345BEZ6 A0A165VTZ5 A0A3S2M0U8 A0A182SPH2 A0A0L7LER6 A0A182RWQ2 A0A182W8Q5 A0A182J5Q6 A0A3B0JCF9 A0A182Y693 A0A182PW40 Q17IE0 A0A182NP16 A0A084VSQ4 A0A1W4V3K1 A0A1S4F1H7 B4Q6J2 M9PAZ2 B5DIW0 B4GPQ5 Q9VPT1 B4ID11 W5J5X2 B4P2X0 A0A0J9QU89 A0A182HR84 Q7PMG3 B3N881 I0E1Y0 A0A1I8MXB7 A0A182X2E9 B3MLR3 A0A182QJA2 A0A1A9ZH76 A0A1A9VXF8 A0A240SWY9 A0A1B3B7E2 A0A1A9W6U1 I0E1X9 A0A1A9XCE1 A0A1B0B0N1 A0A182F548 B4N0S0 A0A1I8MJ63 B4JDS7 A0A0M4EAW5 A0A182GQ98 B0WHB3 A0A1I8NQP4 A0A0L0BUA8 A0A1I8PPJ0 A0A0G2UGF3 A0A348AZU8 B4LTP7 A0A068F7L4 B4KF85 A0A3L9LU10 A0A1J1IQB3 A0A1B0CPT9 A0A336MQ88 A0A310S6C6 A0A0E3Y701 A2AX72 D6WPY0 U4U7B6 N6TZ29 A0A182LX46 A0A2P9JYC0 A0A336N1R5 A0A336MVM5 A0A182K567 A0A223HD96 B0WJE4 A0A0B4ZWF1 A0A075T302 A0A0V0J1N7 A0A3S2LR79 A0A089Q7Y6 A0A223HCV6 A0A2K8GKX0
A0A223HDH7 A0A0V0J1B6 A0A2K8GKX3 A0A0K8TUH7 A0A212ERI7 A0A2H1VDE4 A0A2K8GL76 A0A194RI74 A0A223HD90 A0A194PL61 A0A345BEZ6 A0A165VTZ5 A0A3S2M0U8 A0A182SPH2 A0A0L7LER6 A0A182RWQ2 A0A182W8Q5 A0A182J5Q6 A0A3B0JCF9 A0A182Y693 A0A182PW40 Q17IE0 A0A182NP16 A0A084VSQ4 A0A1W4V3K1 A0A1S4F1H7 B4Q6J2 M9PAZ2 B5DIW0 B4GPQ5 Q9VPT1 B4ID11 W5J5X2 B4P2X0 A0A0J9QU89 A0A182HR84 Q7PMG3 B3N881 I0E1Y0 A0A1I8MXB7 A0A182X2E9 B3MLR3 A0A182QJA2 A0A1A9ZH76 A0A1A9VXF8 A0A240SWY9 A0A1B3B7E2 A0A1A9W6U1 I0E1X9 A0A1A9XCE1 A0A1B0B0N1 A0A182F548 B4N0S0 A0A1I8MJ63 B4JDS7 A0A0M4EAW5 A0A182GQ98 B0WHB3 A0A1I8NQP4 A0A0L0BUA8 A0A1I8PPJ0 A0A0G2UGF3 A0A348AZU8 B4LTP7 A0A068F7L4 B4KF85 A0A3L9LU10 A0A1J1IQB3 A0A1B0CPT9 A0A336MQ88 A0A310S6C6 A0A0E3Y701 A2AX72 D6WPY0 U4U7B6 N6TZ29 A0A182LX46 A0A2P9JYC0 A0A336N1R5 A0A336MVM5 A0A182K567 A0A223HD96 B0WJE4 A0A0B4ZWF1 A0A075T302 A0A0V0J1N7 A0A3S2LR79 A0A089Q7Y6 A0A223HCV6 A0A2K8GKX0
Pubmed
26812239
19121390
25027790
28150741
27006164
28736530
+ More
26017144 22118469 29375398 26354079 29727827 27060445 26227816 25244985 17510324 24438588 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17167414 10710312 11516643 16809465 17493811 17360684 19710651 22031876 23162431 23105004 23185459 20920257 23761445 17550304 22936249 12364791 25315136 27171401 26483478 26108605 30026095 25880816 25856077 18362917 19820115 23537049
26017144 22118469 29375398 26354079 29727827 27060445 26227816 25244985 17510324 24438588 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17167414 10710312 11516643 16809465 17493811 17360684 19710651 22031876 23162431 23105004 23185459 20920257 23761445 17550304 22936249 12364791 25315136 27171401 26483478 26108605 30026095 25880816 25856077 18362917 19820115 23537049
EMBL
KJ542710
AJD81594.1
KR935752
ALM26244.1
BABH01015478
BABH01015479
+ More
LC002742 BAR43490.1 KF768713 AIG51907.1 NWSH01002509 PCG68108.1 KY283762 AST36421.1 GDKB01000001 JAP38495.1 KX585385 ARO70278.1 GCVX01000098 JAI18132.1 AGBW02013022 OWR44079.1 ODYU01001951 SOQ38863.1 KY225526 ARO70538.1 KQ460205 KPJ17139.1 KY283683 AST36342.1 KQ459601 KPI94062.1 MG820649 AXF48820.1 KU664806 AMZ01260.1 RSAL01000081 RVE48553.1 JTDY01001430 KOB73874.1 OUUW01000004 SPP79745.1 CH477240 EAT46439.1 ATLV01016136 KE525057 KFB40998.1 CM000361 EDX03258.1 AE014134 AGB92378.1 CH379060 EDY70253.1 CH479187 EDW39577.1 DQ989014 BT025007 CH480829 EDW45437.1 ADMH02002089 ETN59386.1 CM000157 EDW87177.2 CM002910 KMY87364.1 APCN01001656 DQ989011 AAAB01008980 ABK97612.1 EAA13882.2 CH954177 EDV57268.2 JQ365177 AFH96948.1 CH902620 EDV30784.1 AXCN02001961 CCAG010023722 KU291872 AOE48122.1 JQ365174 AFH96947.1 JXJN01006781 CH963920 EDW77683.1 CH916368 EDW03447.1 CP012523 ALC38447.1 JXUM01015690 KQ560437 KXJ82421.1 DS231934 EDS27619.1 JRES01001455 KNC22794.1 KP743664 AKI28980.1 FX985939 BBD43474.1 CH940649 EDW65020.1 KJ702098 AID61252.1 CH933807 EDW13068.1 KB466104 RLZ02222.1 CVRI01000055 CRL01286.1 AJWK01022645 UFQT01001534 SSX30943.1 GGOB01000037 MCH29433.1 KM251699 AKC58580.2 AM292331 AM292360 CAL23143.2 CAL23172.3 KQ971354 EFA07594.2 KB632188 ERL89794.1 APGK01045842 KB741039 ENN74545.1 AXCM01000378 KY817113 AVH87329.1 UFQS01005150 UFQT01005150 SSX16734.1 SSX35925.1 UFQS01002860 UFQT01002860 SSX14789.1 SSX34180.1 KY283684 AST36343.1 DS231959 EDS29118.1 KJ542711 AJD81595.1 KF768714 AIG51908.1 GDKB01000002 JAP38494.1 RSAL01000025 RVE52125.1 KM056288 AIR07412.1 KY283551 AST36211.1 KX585386 ARO70279.1
LC002742 BAR43490.1 KF768713 AIG51907.1 NWSH01002509 PCG68108.1 KY283762 AST36421.1 GDKB01000001 JAP38495.1 KX585385 ARO70278.1 GCVX01000098 JAI18132.1 AGBW02013022 OWR44079.1 ODYU01001951 SOQ38863.1 KY225526 ARO70538.1 KQ460205 KPJ17139.1 KY283683 AST36342.1 KQ459601 KPI94062.1 MG820649 AXF48820.1 KU664806 AMZ01260.1 RSAL01000081 RVE48553.1 JTDY01001430 KOB73874.1 OUUW01000004 SPP79745.1 CH477240 EAT46439.1 ATLV01016136 KE525057 KFB40998.1 CM000361 EDX03258.1 AE014134 AGB92378.1 CH379060 EDY70253.1 CH479187 EDW39577.1 DQ989014 BT025007 CH480829 EDW45437.1 ADMH02002089 ETN59386.1 CM000157 EDW87177.2 CM002910 KMY87364.1 APCN01001656 DQ989011 AAAB01008980 ABK97612.1 EAA13882.2 CH954177 EDV57268.2 JQ365177 AFH96948.1 CH902620 EDV30784.1 AXCN02001961 CCAG010023722 KU291872 AOE48122.1 JQ365174 AFH96947.1 JXJN01006781 CH963920 EDW77683.1 CH916368 EDW03447.1 CP012523 ALC38447.1 JXUM01015690 KQ560437 KXJ82421.1 DS231934 EDS27619.1 JRES01001455 KNC22794.1 KP743664 AKI28980.1 FX985939 BBD43474.1 CH940649 EDW65020.1 KJ702098 AID61252.1 CH933807 EDW13068.1 KB466104 RLZ02222.1 CVRI01000055 CRL01286.1 AJWK01022645 UFQT01001534 SSX30943.1 GGOB01000037 MCH29433.1 KM251699 AKC58580.2 AM292331 AM292360 CAL23143.2 CAL23172.3 KQ971354 EFA07594.2 KB632188 ERL89794.1 APGK01045842 KB741039 ENN74545.1 AXCM01000378 KY817113 AVH87329.1 UFQS01005150 UFQT01005150 SSX16734.1 SSX35925.1 UFQS01002860 UFQT01002860 SSX14789.1 SSX34180.1 KY283684 AST36343.1 DS231959 EDS29118.1 KJ542711 AJD81595.1 KF768714 AIG51908.1 GDKB01000002 JAP38494.1 RSAL01000025 RVE52125.1 KM056288 AIR07412.1 KY283551 AST36211.1 KX585386 ARO70279.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000075901 UP000037510 UP000075900 UP000075920 UP000075880 UP000268350 UP000076408 UP000075885 UP000008820 UP000075884 UP000030765 UP000192221 UP000000304 UP000000803 UP000001819 UP000008744 UP000001292 UP000000673 UP000002282 UP000075840 UP000007062 UP000008711 UP000095301 UP000076407 UP000007801 UP000075886 UP000092445 UP000078200 UP000092444 UP000091820 UP000092443 UP000092460 UP000069272 UP000007798 UP000001070 UP000092553 UP000069940 UP000249989 UP000002320 UP000095300 UP000037069 UP000008792 UP000009192 UP000183832 UP000092461 UP000007266 UP000030742 UP000019118 UP000075883 UP000075881
UP000075901 UP000037510 UP000075900 UP000075920 UP000075880 UP000268350 UP000076408 UP000075885 UP000008820 UP000075884 UP000030765 UP000192221 UP000000304 UP000000803 UP000001819 UP000008744 UP000001292 UP000000673 UP000002282 UP000075840 UP000007062 UP000008711 UP000095301 UP000076407 UP000007801 UP000075886 UP000092445 UP000078200 UP000092444 UP000091820 UP000092443 UP000092460 UP000069272 UP000007798 UP000001070 UP000092553 UP000069940 UP000249989 UP000002320 UP000095300 UP000037069 UP000008792 UP000009192 UP000183832 UP000092461 UP000007266 UP000030742 UP000019118 UP000075883 UP000075881
Pfam
PF08395 7tm_7
Interpro
IPR013604
7TM_chemorcpt
ProteinModelPortal
A0A0B5A3E3
A0A0S1TPL0
H9JPX4
A0A0E4B436
A0A075T806
A0A2A4J8U7
+ More
A0A223HDH7 A0A0V0J1B6 A0A2K8GKX3 A0A0K8TUH7 A0A212ERI7 A0A2H1VDE4 A0A2K8GL76 A0A194RI74 A0A223HD90 A0A194PL61 A0A345BEZ6 A0A165VTZ5 A0A3S2M0U8 A0A182SPH2 A0A0L7LER6 A0A182RWQ2 A0A182W8Q5 A0A182J5Q6 A0A3B0JCF9 A0A182Y693 A0A182PW40 Q17IE0 A0A182NP16 A0A084VSQ4 A0A1W4V3K1 A0A1S4F1H7 B4Q6J2 M9PAZ2 B5DIW0 B4GPQ5 Q9VPT1 B4ID11 W5J5X2 B4P2X0 A0A0J9QU89 A0A182HR84 Q7PMG3 B3N881 I0E1Y0 A0A1I8MXB7 A0A182X2E9 B3MLR3 A0A182QJA2 A0A1A9ZH76 A0A1A9VXF8 A0A240SWY9 A0A1B3B7E2 A0A1A9W6U1 I0E1X9 A0A1A9XCE1 A0A1B0B0N1 A0A182F548 B4N0S0 A0A1I8MJ63 B4JDS7 A0A0M4EAW5 A0A182GQ98 B0WHB3 A0A1I8NQP4 A0A0L0BUA8 A0A1I8PPJ0 A0A0G2UGF3 A0A348AZU8 B4LTP7 A0A068F7L4 B4KF85 A0A3L9LU10 A0A1J1IQB3 A0A1B0CPT9 A0A336MQ88 A0A310S6C6 A0A0E3Y701 A2AX72 D6WPY0 U4U7B6 N6TZ29 A0A182LX46 A0A2P9JYC0 A0A336N1R5 A0A336MVM5 A0A182K567 A0A223HD96 B0WJE4 A0A0B4ZWF1 A0A075T302 A0A0V0J1N7 A0A3S2LR79 A0A089Q7Y6 A0A223HCV6 A0A2K8GKX0
A0A223HDH7 A0A0V0J1B6 A0A2K8GKX3 A0A0K8TUH7 A0A212ERI7 A0A2H1VDE4 A0A2K8GL76 A0A194RI74 A0A223HD90 A0A194PL61 A0A345BEZ6 A0A165VTZ5 A0A3S2M0U8 A0A182SPH2 A0A0L7LER6 A0A182RWQ2 A0A182W8Q5 A0A182J5Q6 A0A3B0JCF9 A0A182Y693 A0A182PW40 Q17IE0 A0A182NP16 A0A084VSQ4 A0A1W4V3K1 A0A1S4F1H7 B4Q6J2 M9PAZ2 B5DIW0 B4GPQ5 Q9VPT1 B4ID11 W5J5X2 B4P2X0 A0A0J9QU89 A0A182HR84 Q7PMG3 B3N881 I0E1Y0 A0A1I8MXB7 A0A182X2E9 B3MLR3 A0A182QJA2 A0A1A9ZH76 A0A1A9VXF8 A0A240SWY9 A0A1B3B7E2 A0A1A9W6U1 I0E1X9 A0A1A9XCE1 A0A1B0B0N1 A0A182F548 B4N0S0 A0A1I8MJ63 B4JDS7 A0A0M4EAW5 A0A182GQ98 B0WHB3 A0A1I8NQP4 A0A0L0BUA8 A0A1I8PPJ0 A0A0G2UGF3 A0A348AZU8 B4LTP7 A0A068F7L4 B4KF85 A0A3L9LU10 A0A1J1IQB3 A0A1B0CPT9 A0A336MQ88 A0A310S6C6 A0A0E3Y701 A2AX72 D6WPY0 U4U7B6 N6TZ29 A0A182LX46 A0A2P9JYC0 A0A336N1R5 A0A336MVM5 A0A182K567 A0A223HD96 B0WJE4 A0A0B4ZWF1 A0A075T302 A0A0V0J1N7 A0A3S2LR79 A0A089Q7Y6 A0A223HCV6 A0A2K8GKX0
Ontologies
GO
PANTHER
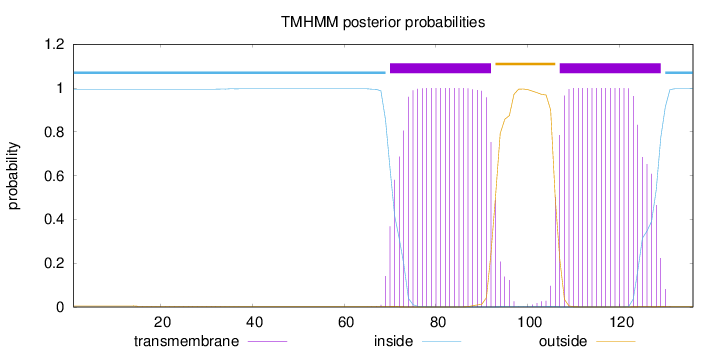
Topology
Subcellular location
Cell membrane
Length:
136
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.18672
Exp number, first 60 AAs:
0.03606
Total prob of N-in:
0.99492
inside
1 - 69
TMhelix
70 - 92
outside
93 - 106
TMhelix
107 - 129
inside
130 - 136
Population Genetic Test Statistics
Pi
228.099411
Theta
188.24795
Tajima's D
0.670482
CLR
0
CSRT
0.567171641417929
Interpretation
Uncertain
