Pre Gene Modal
BGIBMGA011577
Annotation
Monocarboxylate_transporter_12-B_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.864
Sequence
CDS
ATGGTGGTGAGGGTAGCAACAGATGAGCGGCCTTTTGAGAGTCCTTCTTCGGATGAGAGTGGGCTCGGGAGAAGCGACTCTCCCAGCGAAAACACTGAACCAGAAGCAGCATTGGTGGTTCCTCCAGATGGAGGCTGGGGCTGGGTTGTGGTGGGAGCGTCTTTCATGTGCAATGTATTAGTTGATGGCATCATATTCTCTGGAGGTCTGTTGTCCTCTGCCATCAAGGATGATTTTAAGATTAGTGAAGGAAAAGTAGCTACTGTGAATTCTCTACTGGCTGGCTTCTACTTATTGGCTGGACCTTTCGTGTCGGCACTAGCTAACAAGTATGGATTTCGCGTCGTAACAATAATCGGAAGTTTGATTTCGAGTTTTGCATTCGCTGTCTCCTACTACGCAACCAGCGTAGAATATCTTTATATCATCTACGGTATAATGGGAGGTATTGGACTATGTATGATATATATGCCTGCGGTGCTGACTGTTGGTTTCTACTTTGAAAAGTGGAGAGGTCTAGCCACTGGCTTAGCACTTTGTGGTTCCGGAGTAGGCACTTTCGTTATCGCCATCTTAACTAATAAACTAAATGAAAGCTTACAAAATTGGAGACTTATTGTGCTCATACATGCAGCCATGTTACTCCTATGCATGCTCTTTGGTGCTCTATTCCGGCCGATCAAACCAGTCCGCGTGACATTAGCCGACGAACAAGATGAAGAAGATACTGCAAGACGTCACGAGGAGGCCGTCGAAAAACTTAACTCCATGATCAAGTTACACAGCAAACTCGATTCTGGAATTTGCATGCCTCCAGAAATGAAGTTTACAAACAAAGTAAGCCCTCATACTTGGATGGGCGTGCCTAATAACACACGTTACCCGACTGCCGAAGAGGTATTCCGTGGCAGTAACTCGCATTTGAGCAATTCACACGCCGGGAGACGATCGTCAGCGAATGCTGGAGCTATAAAGAATACACTTGGAAACAAACCTATGTTCATCGAAATGCCAGTCGCTGAGAAAGATGAACAAGAGGATTCAAACGGTAACATCGACAACACTGAGCCCCTTATTGGAGCTGCTGTGAAAGTTGTCCCCACACAGAACCGTTTCCATCAACGTCGTTCTCATGCTGATTTAGTGGCCAGACCCTTGTACCGGGACGATATTTTCTTTGGAGCCAGTTTGACGCGATTGCCTCAATATACATCTAGGACTTCGCTTGGATACCATCTCGCAGTAACGCACGTTCCTACAAAAGAAGATGCTCAAGAGGAATCCTCAGGAAAATGTCGTTTCTGCCCTGAAGCCGTGAAAAGGACATTGGCTACCATGTTAGACGTCAGTCTATTCCGATCACCGACCTTTGTCATCCTTGCTCTAAGTGGATTTTTCACAATGTTAGGCTTCTTCGTTCCGTACATGTACATACAGAGACGAGGTGAAAGTAACGGTATGAATAAAGCAATCGCCGGTTGGCTTATATCCGCCATAGGAATATCGAACATAGTTGGTCGTGTGATGTGTGGATTTCTATCGTCGATGCCGAAGATATCCCCTTTGTGGTTGAACAACATCGCCATCTCCGCCGGGGGGATTGCAACTATGATGAGCGGACTGTACTACGACAGCGTTTTCCAATTTGCCTATTGCGCCATTTTCGGACTGGCTGCTGCGTGCTTCGCCTCTCTCCGTTCAATTTTGGTAGTGGAGTATATTGGACTAGAACAGTTAACCAACTGTTTCGGGCTATTCCTCCTGTTCCAAGGGTTTGGTGCTGTGTTGGGAGTGCCTATTGCAGGCATGCTCTATGACCTGACCCTTAGTTACGATAAATCGTTCTACGTGTCCGGTGCATTCCTGTTGTTGTCTGCGGTTATGTGTTATCCTGTGGATGCGATCAGCAAATGGGAGAAGTCTCGCAAAGTACACAACTCTTCGCCGACAGCTTGA
Protein
MVVRVATDERPFESPSSDESGLGRSDSPSENTEPEAALVVPPDGGWGWVVVGASFMCNVLVDGIIFSGGLLSSAIKDDFKISEGKVATVNSLLAGFYLLAGPFVSALANKYGFRVVTIIGSLISSFAFAVSYYATSVEYLYIIYGIMGGIGLCMIYMPAVLTVGFYFEKWRGLATGLALCGSGVGTFVIAILTNKLNESLQNWRLIVLIHAAMLLLCMLFGALFRPIKPVRVTLADEQDEEDTARRHEEAVEKLNSMIKLHSKLDSGICMPPEMKFTNKVSPHTWMGVPNNTRYPTAEEVFRGSNSHLSNSHAGRRSSANAGAIKNTLGNKPMFIEMPVAEKDEQEDSNGNIDNTEPLIGAAVKVVPTQNRFHQRRSHADLVARPLYRDDIFFGASLTRLPQYTSRTSLGYHLAVTHVPTKEDAQEESSGKCRFCPEAVKRTLATMLDVSLFRSPTFVILALSGFFTMLGFFVPYMYIQRRGESNGMNKAIAGWLISAIGISNIVGRVMCGFLSSMPKISPLWLNNIAISAGGIATMMSGLYYDSVFQFAYCAIFGLAAACFASLRSILVVEYIGLEQLTNCFGLFLLFQGFGAVLGVPIAGMLYDLTLSYDKSFYVSGAFLLLSAVMCYPVDAISKWEKSRKVHNSSPTA
Summary
Uniprot
H9JPX2
A0A212EXA5
A0A3S2TH11
A0A194RMB6
A0A194PKU4
A0A0L7KWQ8
+ More
A0A0L7K4P1 A0A2A4JHP8 Q17P95 A0A182GZ05 A0A1S4EVY2 A0A1B0GL20 A0A1L8DEJ8 B0W6P3 A0A1Q3F8J6 A0A182QYV6 A0A182WQN9 A0A182LUI9 A0A182S8G5 A0A182ISG7 Q7PGV2 A0A084WSJ3 A0A182URT2 A0A182TNZ5 A0A0T6BDX1 U5EE52 A0A2M4ADU5 W5J6W5 A0A182N5C1 A0A1Y1KD00 A0A2M4BFY8 A0A182JYR7 A0A182XVZ9 A0A182PE32 A0A0P4VR86 A0A0L0CB43 A0A069DWI1 A0A1B0AM52 T1HU50 A0A224X8K5 A0A1A9XPS1 A0A1B0GC16 A0A088A5V0 A0A0M4EDR0 W8ASF1 A0A2J7QT03 B4P6N9 A0A1A9V7P5 A0A023F0W6 A0A182HJ87 A0A1A9WNC8 Q7K1L4 A0A0A1WYG7 A0A0J9RCG7 B3MCY4 B4J7M6 B3NR74 A0A1W4VB53 A0A1A9ZPB5 B4QFE3 A0A1W4X5L6 A0A1W4XG10 A0A1W4X5Q0 E9J9H6 A0A1S3DA16 A0A3Q0J3X9 A0A0K8VGG0 B4MPQ6 A0A0R3NQ65 Q28X65 E1ZY62 D6WA62 A0A0L7R1S2 K7IRA1 B4H530 B4KRC0 A0A3B0JMY1 A0A182RIV7 A0A195BET9 A0A158NBY3 T1PDW7 B4LLD9 A0A151IV39 A0A3L8DSX0 F4WHW2 E2BUR3 A0A182XE36 A0A195FM12 A0A026VVK5 A0A146M3F9 A0A0A9WFY8 A0A1L8EFV5 A0A1B6DPH3 A0A1I8PXA0 B4HR28 A0A182FRW9 A0A1B6FNF8 A0A1B6JCU5
A0A0L7K4P1 A0A2A4JHP8 Q17P95 A0A182GZ05 A0A1S4EVY2 A0A1B0GL20 A0A1L8DEJ8 B0W6P3 A0A1Q3F8J6 A0A182QYV6 A0A182WQN9 A0A182LUI9 A0A182S8G5 A0A182ISG7 Q7PGV2 A0A084WSJ3 A0A182URT2 A0A182TNZ5 A0A0T6BDX1 U5EE52 A0A2M4ADU5 W5J6W5 A0A182N5C1 A0A1Y1KD00 A0A2M4BFY8 A0A182JYR7 A0A182XVZ9 A0A182PE32 A0A0P4VR86 A0A0L0CB43 A0A069DWI1 A0A1B0AM52 T1HU50 A0A224X8K5 A0A1A9XPS1 A0A1B0GC16 A0A088A5V0 A0A0M4EDR0 W8ASF1 A0A2J7QT03 B4P6N9 A0A1A9V7P5 A0A023F0W6 A0A182HJ87 A0A1A9WNC8 Q7K1L4 A0A0A1WYG7 A0A0J9RCG7 B3MCY4 B4J7M6 B3NR74 A0A1W4VB53 A0A1A9ZPB5 B4QFE3 A0A1W4X5L6 A0A1W4XG10 A0A1W4X5Q0 E9J9H6 A0A1S3DA16 A0A3Q0J3X9 A0A0K8VGG0 B4MPQ6 A0A0R3NQ65 Q28X65 E1ZY62 D6WA62 A0A0L7R1S2 K7IRA1 B4H530 B4KRC0 A0A3B0JMY1 A0A182RIV7 A0A195BET9 A0A158NBY3 T1PDW7 B4LLD9 A0A151IV39 A0A3L8DSX0 F4WHW2 E2BUR3 A0A182XE36 A0A195FM12 A0A026VVK5 A0A146M3F9 A0A0A9WFY8 A0A1L8EFV5 A0A1B6DPH3 A0A1I8PXA0 B4HR28 A0A182FRW9 A0A1B6FNF8 A0A1B6JCU5
Pubmed
19121390
22118469
26354079
26227816
17510324
26483478
+ More
12364791 14747013 17210077 24438588 20920257 23761445 28004739 25244985 27129103 26108605 26334808 24495485 17994087 17550304 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 21282665 15632085 20798317 18362917 19820115 20075255 21347285 25315136 30249741 21719571 24508170 26823975 25401762
12364791 14747013 17210077 24438588 20920257 23761445 28004739 25244985 27129103 26108605 26334808 24495485 17994087 17550304 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 21282665 15632085 20798317 18362917 19820115 20075255 21347285 25315136 30249741 21719571 24508170 26823975 25401762
EMBL
BABH01015452
BABH01015453
AGBW02011818
OWR46101.1
RSAL01000149
RVE45812.1
+ More
KQ460205 KPJ17131.1 KQ459601 KPI94056.1 JTDY01004856 KOB67673.1 JTDY01010483 KOB58187.1 NWSH01001527 PCG70963.1 CH477193 EAT48537.1 JXUM01021827 JXUM01021828 JXUM01021829 JXUM01021830 JXUM01021831 KQ560612 KXJ81625.1 AJWK01034066 AJWK01034067 AJWK01034068 AJWK01034069 AJWK01034070 GFDF01009226 JAV04858.1 DS231849 EDS36874.1 GFDL01011179 JAV23866.1 AXCN02000147 AXCN02000148 AXCM01002064 AAAB01008879 EAA44794.4 ATLV01026580 KE525415 KFB53187.1 LJIG01001502 KRT85438.1 GANO01004451 JAB55420.1 GGFK01005628 MBW38949.1 ADMH02002129 ETN58605.1 GEZM01088608 JAV58070.1 GGFJ01002813 MBW51954.1 GDKW01001540 JAI55055.1 JRES01000644 KNC29638.1 GBGD01000688 JAC88201.1 JXJN01000337 ACPB03003092 GFTR01007760 JAW08666.1 CCAG010018970 CCAG010018971 CCAG010018972 CP012524 ALC41164.1 GAMC01017528 GAMC01017527 JAB89027.1 NEVH01011202 PNF31722.1 CM000158 EDW90991.2 KRJ99554.1 GBBI01004056 JAC14656.1 AE013599 AY061626 AAF58276.1 AAL29174.1 GBXI01013914 GBXI01010844 GBXI01008820 JAD00378.1 JAD03448.1 JAD05472.1 CM002911 KMY93723.1 CH902619 EDV36299.1 CH916367 EDW02174.1 CH954179 EDV56063.1 CM000362 EDX07053.1 GL769313 EFZ10530.1 GDHF01028570 GDHF01027445 GDHF01014305 GDHF01011196 GDHF01004681 JAI23744.1 JAI24869.1 JAI38009.1 JAI41118.1 JAI47633.1 CH963849 EDW74095.1 CM000071 KRT03114.1 EAL26451.2 GL435171 EFN73911.1 KQ971312 EEZ98054.2 KQ414667 KOC64807.1 CH479210 EDW32866.1 CH933808 EDW09336.1 OUUW01000001 SPP74676.1 KQ976500 KYM83088.1 ADTU01011482 ADTU01011483 ADTU01011484 ADTU01011485 KA646936 AFP61565.1 CH940648 EDW61891.1 KQ980949 KYN11209.1 QOIP01000004 RLU23535.1 GL888170 EGI66191.1 GL450743 EFN80568.1 KQ981491 KYN41024.1 KK107785 EZA47787.1 GDHC01012090 GDHC01004660 JAQ06539.1 JAQ13969.1 GBHO01039830 GBHO01039829 JAG03774.1 JAG03775.1 GFDG01001218 JAV17581.1 GEDC01009712 JAS27586.1 CH480816 EDW47825.1 GECZ01018022 JAS51747.1 GECU01033523 GECU01027355 GECU01026512 GECU01019257 GECU01010585 GECU01007195 JAS74183.1 JAS80351.1 JAS81194.1 JAS88449.1 JAS97121.1 JAT00512.1
KQ460205 KPJ17131.1 KQ459601 KPI94056.1 JTDY01004856 KOB67673.1 JTDY01010483 KOB58187.1 NWSH01001527 PCG70963.1 CH477193 EAT48537.1 JXUM01021827 JXUM01021828 JXUM01021829 JXUM01021830 JXUM01021831 KQ560612 KXJ81625.1 AJWK01034066 AJWK01034067 AJWK01034068 AJWK01034069 AJWK01034070 GFDF01009226 JAV04858.1 DS231849 EDS36874.1 GFDL01011179 JAV23866.1 AXCN02000147 AXCN02000148 AXCM01002064 AAAB01008879 EAA44794.4 ATLV01026580 KE525415 KFB53187.1 LJIG01001502 KRT85438.1 GANO01004451 JAB55420.1 GGFK01005628 MBW38949.1 ADMH02002129 ETN58605.1 GEZM01088608 JAV58070.1 GGFJ01002813 MBW51954.1 GDKW01001540 JAI55055.1 JRES01000644 KNC29638.1 GBGD01000688 JAC88201.1 JXJN01000337 ACPB03003092 GFTR01007760 JAW08666.1 CCAG010018970 CCAG010018971 CCAG010018972 CP012524 ALC41164.1 GAMC01017528 GAMC01017527 JAB89027.1 NEVH01011202 PNF31722.1 CM000158 EDW90991.2 KRJ99554.1 GBBI01004056 JAC14656.1 AE013599 AY061626 AAF58276.1 AAL29174.1 GBXI01013914 GBXI01010844 GBXI01008820 JAD00378.1 JAD03448.1 JAD05472.1 CM002911 KMY93723.1 CH902619 EDV36299.1 CH916367 EDW02174.1 CH954179 EDV56063.1 CM000362 EDX07053.1 GL769313 EFZ10530.1 GDHF01028570 GDHF01027445 GDHF01014305 GDHF01011196 GDHF01004681 JAI23744.1 JAI24869.1 JAI38009.1 JAI41118.1 JAI47633.1 CH963849 EDW74095.1 CM000071 KRT03114.1 EAL26451.2 GL435171 EFN73911.1 KQ971312 EEZ98054.2 KQ414667 KOC64807.1 CH479210 EDW32866.1 CH933808 EDW09336.1 OUUW01000001 SPP74676.1 KQ976500 KYM83088.1 ADTU01011482 ADTU01011483 ADTU01011484 ADTU01011485 KA646936 AFP61565.1 CH940648 EDW61891.1 KQ980949 KYN11209.1 QOIP01000004 RLU23535.1 GL888170 EGI66191.1 GL450743 EFN80568.1 KQ981491 KYN41024.1 KK107785 EZA47787.1 GDHC01012090 GDHC01004660 JAQ06539.1 JAQ13969.1 GBHO01039830 GBHO01039829 JAG03774.1 JAG03775.1 GFDG01001218 JAV17581.1 GEDC01009712 JAS27586.1 CH480816 EDW47825.1 GECZ01018022 JAS51747.1 GECU01033523 GECU01027355 GECU01026512 GECU01019257 GECU01010585 GECU01007195 JAS74183.1 JAS80351.1 JAS81194.1 JAS88449.1 JAS97121.1 JAT00512.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000218220 UP000008820 UP000069940 UP000249989 UP000092461 UP000002320 UP000075886 UP000075920 UP000075883 UP000075901 UP000075880 UP000007062 UP000030765 UP000075903 UP000075902 UP000000673 UP000075884 UP000075881 UP000076408 UP000075885 UP000037069 UP000092460 UP000015103 UP000092443 UP000092444 UP000005203 UP000092553 UP000235965 UP000002282 UP000078200 UP000091820 UP000000803 UP000007801 UP000001070 UP000008711 UP000192221 UP000092445 UP000000304 UP000192223 UP000079169 UP000007798 UP000001819 UP000000311 UP000007266 UP000053825 UP000002358 UP000008744 UP000009192 UP000268350 UP000075900 UP000078540 UP000005205 UP000095301 UP000008792 UP000078492 UP000279307 UP000007755 UP000008237 UP000076407 UP000078541 UP000053097 UP000095300 UP000001292 UP000069272
UP000218220 UP000008820 UP000069940 UP000249989 UP000092461 UP000002320 UP000075886 UP000075920 UP000075883 UP000075901 UP000075880 UP000007062 UP000030765 UP000075903 UP000075902 UP000000673 UP000075884 UP000075881 UP000076408 UP000075885 UP000037069 UP000092460 UP000015103 UP000092443 UP000092444 UP000005203 UP000092553 UP000235965 UP000002282 UP000078200 UP000091820 UP000000803 UP000007801 UP000001070 UP000008711 UP000192221 UP000092445 UP000000304 UP000192223 UP000079169 UP000007798 UP000001819 UP000000311 UP000007266 UP000053825 UP000002358 UP000008744 UP000009192 UP000268350 UP000075900 UP000078540 UP000005205 UP000095301 UP000008792 UP000078492 UP000279307 UP000007755 UP000008237 UP000076407 UP000078541 UP000053097 UP000095300 UP000001292 UP000069272
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JPX2
A0A212EXA5
A0A3S2TH11
A0A194RMB6
A0A194PKU4
A0A0L7KWQ8
+ More
A0A0L7K4P1 A0A2A4JHP8 Q17P95 A0A182GZ05 A0A1S4EVY2 A0A1B0GL20 A0A1L8DEJ8 B0W6P3 A0A1Q3F8J6 A0A182QYV6 A0A182WQN9 A0A182LUI9 A0A182S8G5 A0A182ISG7 Q7PGV2 A0A084WSJ3 A0A182URT2 A0A182TNZ5 A0A0T6BDX1 U5EE52 A0A2M4ADU5 W5J6W5 A0A182N5C1 A0A1Y1KD00 A0A2M4BFY8 A0A182JYR7 A0A182XVZ9 A0A182PE32 A0A0P4VR86 A0A0L0CB43 A0A069DWI1 A0A1B0AM52 T1HU50 A0A224X8K5 A0A1A9XPS1 A0A1B0GC16 A0A088A5V0 A0A0M4EDR0 W8ASF1 A0A2J7QT03 B4P6N9 A0A1A9V7P5 A0A023F0W6 A0A182HJ87 A0A1A9WNC8 Q7K1L4 A0A0A1WYG7 A0A0J9RCG7 B3MCY4 B4J7M6 B3NR74 A0A1W4VB53 A0A1A9ZPB5 B4QFE3 A0A1W4X5L6 A0A1W4XG10 A0A1W4X5Q0 E9J9H6 A0A1S3DA16 A0A3Q0J3X9 A0A0K8VGG0 B4MPQ6 A0A0R3NQ65 Q28X65 E1ZY62 D6WA62 A0A0L7R1S2 K7IRA1 B4H530 B4KRC0 A0A3B0JMY1 A0A182RIV7 A0A195BET9 A0A158NBY3 T1PDW7 B4LLD9 A0A151IV39 A0A3L8DSX0 F4WHW2 E2BUR3 A0A182XE36 A0A195FM12 A0A026VVK5 A0A146M3F9 A0A0A9WFY8 A0A1L8EFV5 A0A1B6DPH3 A0A1I8PXA0 B4HR28 A0A182FRW9 A0A1B6FNF8 A0A1B6JCU5
A0A0L7K4P1 A0A2A4JHP8 Q17P95 A0A182GZ05 A0A1S4EVY2 A0A1B0GL20 A0A1L8DEJ8 B0W6P3 A0A1Q3F8J6 A0A182QYV6 A0A182WQN9 A0A182LUI9 A0A182S8G5 A0A182ISG7 Q7PGV2 A0A084WSJ3 A0A182URT2 A0A182TNZ5 A0A0T6BDX1 U5EE52 A0A2M4ADU5 W5J6W5 A0A182N5C1 A0A1Y1KD00 A0A2M4BFY8 A0A182JYR7 A0A182XVZ9 A0A182PE32 A0A0P4VR86 A0A0L0CB43 A0A069DWI1 A0A1B0AM52 T1HU50 A0A224X8K5 A0A1A9XPS1 A0A1B0GC16 A0A088A5V0 A0A0M4EDR0 W8ASF1 A0A2J7QT03 B4P6N9 A0A1A9V7P5 A0A023F0W6 A0A182HJ87 A0A1A9WNC8 Q7K1L4 A0A0A1WYG7 A0A0J9RCG7 B3MCY4 B4J7M6 B3NR74 A0A1W4VB53 A0A1A9ZPB5 B4QFE3 A0A1W4X5L6 A0A1W4XG10 A0A1W4X5Q0 E9J9H6 A0A1S3DA16 A0A3Q0J3X9 A0A0K8VGG0 B4MPQ6 A0A0R3NQ65 Q28X65 E1ZY62 D6WA62 A0A0L7R1S2 K7IRA1 B4H530 B4KRC0 A0A3B0JMY1 A0A182RIV7 A0A195BET9 A0A158NBY3 T1PDW7 B4LLD9 A0A151IV39 A0A3L8DSX0 F4WHW2 E2BUR3 A0A182XE36 A0A195FM12 A0A026VVK5 A0A146M3F9 A0A0A9WFY8 A0A1L8EFV5 A0A1B6DPH3 A0A1I8PXA0 B4HR28 A0A182FRW9 A0A1B6FNF8 A0A1B6JCU5
Ontologies
GO
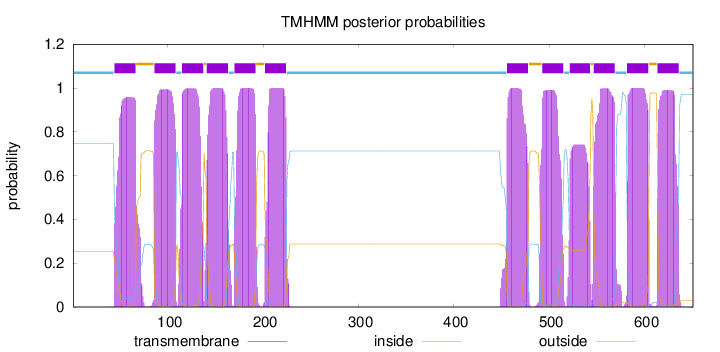
Topology
Length:
651
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
258.79678
Exp number, first 60 AAs:
13.96443
Total prob of N-in:
0.74654
POSSIBLE N-term signal
sequence
inside
1 - 43
TMhelix
44 - 66
outside
67 - 85
TMhelix
86 - 108
inside
109 - 114
TMhelix
115 - 137
outside
138 - 140
TMhelix
141 - 163
inside
164 - 169
TMhelix
170 - 192
outside
193 - 201
TMhelix
202 - 224
inside
225 - 455
TMhelix
456 - 478
outside
479 - 492
TMhelix
493 - 515
inside
516 - 521
TMhelix
522 - 543
outside
544 - 546
TMhelix
547 - 569
inside
570 - 581
TMhelix
582 - 604
outside
605 - 613
TMhelix
614 - 636
inside
637 - 651
Population Genetic Test Statistics
Pi
241.600756
Theta
190.949297
Tajima's D
0.819766
CLR
0.105774
CSRT
0.615219239038048
Interpretation
Uncertain
