Gene
KWMTBOMO13856
Pre Gene Modal
BGIBMGA011576
Annotation
PREDICTED:_monocarboxylate_transporter_4_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.622
Sequence
CDS
ATGTCAAAGGTGTCGCTGAATAACGTTCCATCGAACCCTCGTATTAGTCGTCTCAGGACCTTCTCCAGGACGGAAAGCGAAACCTCATGTGAAGAGGCTGCAAGACTGACAGCGGAAGGAGCGGAAGCCGACGACGATGAGGCCGTCGACTTTACAGCCCTGCCCCCACCGCCGGACGGAGGCTACGGCTGGGTCGTGGTCTTCGCTTCCTTCATGTGCAATATGGTAGTTGACGGCATAGCCTATACCTTTGGCATATTCCTGCCGGAATTGGTGGTGTACTTTGGCCAGGGCAAAGGGACAGTCGCATGGGTCGGCAGTTTGCTGACCGGTATTTATTTAGCCGTTGGTCCGATTGTGTCGGCGTTATGCAACAAATATGGTTGTCGAGCGGTTTGCATCGTGGGCTCCCTAATCGCTACTGCTGCATTCATAGCTTCAACGTTCAGCAAGAGCGTTCCTATGATGATGTTGACGTACGGACTCATCGGTGGGATCGGTTTCGGTATGATCTACCTGCCGTCGGTTGTGGCAGTGGGGTATTACTTCGAAGCGCGTCGTTCGTTGGCGACCGGAATAGCGGTGTGCGGCTCGGGCGTAGGCACATTCGGGTTTTCGCCCCTCGCCACCGCTCTGCTGCACGAGTTCGGTACATGGCAGGGCGCGAACCTCCTGCTGGCCGGTCTCATATTAAACTGCGCTATATTTGGCGCTTTAATGAGACCCCTCGTTTATCCTAAGACTTCAGGCGAGAAACCCCTTCTGCAACGTATGGCCGAAGAGAAGAGACTGCAGATGGAACGGGGGTCCATAGGCGGTTCGTATTTCGTGGTGCAGCTTCCCGACGGCACCATGGAGAAGAGACTAAAGGCCCCATTGAACATCGATCCCGGAGTACATTCGTCGTTGAACCTGGAGGCTCTGGCCCGGGTGCCCACCGTGCCCAACATGCACGGCGTGCCCACAGTGCCCACTTTGCCCACAATCACTGAGGCTAAAGTCGTCGACACTACCGACAAAAAGAAGAACGAGAACGGGAACGTTATAGTGAACCCCGTGCAAGTGCAAATATCGCGCAACGTGTCGTCGCCCGCGTTCAGCGCGCAGGCCCCCGGGCTGCCCAAGAACGGCTCGGTCCCGTTCTTCGACAGGCAGCGCAAGCACAGCACGGGCGAGAAAGGTGCTGAAAGATTCAAGCCGTCCCTAGCAGCCATCAAAGCGACTTCGAAGACCTCCGTCGGCAGCCACCGACCGACGCCAGATGGCGACACAGAAAGCAACATGTACAACTCTAAGTTGTCGGTCTCTGGGCCAAGGGAAGCGCCCCGCATGGTCCGACCGATGTCCCGTAAAGACATCTTCTACTCCGGTTCGGTGCTCAATCTACCTCAGTACCAGAGTCAGAAGTCTCTACAAGGATACAGGAATTCCGTGCTCAGCCTTCCACAGAGCAGGCAGACCGGGGACCTGGAACGACAAGAACAATACGACCTCTGTCCTTGCCTTCACTTGCCGAATTCGTTCAAAAGCGCGTTGTCTTCAATGCTGGATGTAAGCCTGCTGAAGGACCCCGCCTTCATGCTCATCGGCGTCTCCAACATATTCGGGATGGCCGGACTGTACGTGCCCTTCGTGTACCTCACGGACGCAGCCAAAGTTAACGGCATAGAGCCGTCGCAAGCATCGTTCCTGCTGTCCATAATCGGTATAACGAATACGTTCGGCCGTATCATATGCGGGTGGGTCGCAGACTTTCCCTGGATGGACTCCTTGATGCTGAACAACGTGTGCCTGGTCATTGCCACGGTTTCTGTCGGTGTAACGCCCCTGTGCACCACGTACAGTGCCTACGTAGTGATTGCTATAACTTTTGGGATTGCTATTTCTGGCTATATCTCGCTGACGTCGATCATCCTGGTGGATTTGCTGGGGCTGGAGAAGCTGACGAACGCGTTCGGGCTGCTGATCCTGTTCCGCGGCGCCGCGGCCATGGTGGGCTCCCCGCTGGCCGGCGCCGTGTTCGACGCCACGGGGACTTACGGACCCTCCTTCTACATGGCCTCCGTTTTCTTCCTGATATCGACCTTGACCTCGTTCGCCGCACCCTTGTTCCGCAGAAAACAAGAGGAGATACCGCAGCCGTTAGACGTTTTGACTCCCATAGACGAGGATCTAGAGGAGGGTGACGAGGAGGACCCCGACGACACTCCCATCACGATGGGAGCGCAACTGGCGAGCCGCACTCCCGCCATCACCCGCACTGCGGCCTCTCCCTCCGACCCTCCCTCCCCTACAAACAACGAACTGCAGCAAAGGGAAAGCGTGCTCTAG
Protein
MSKVSLNNVPSNPRISRLRTFSRTESETSCEEAARLTAEGAEADDDEAVDFTALPPPPDGGYGWVVVFASFMCNMVVDGIAYTFGIFLPELVVYFGQGKGTVAWVGSLLTGIYLAVGPIVSALCNKYGCRAVCIVGSLIATAAFIASTFSKSVPMMMLTYGLIGGIGFGMIYLPSVVAVGYYFEARRSLATGIAVCGSGVGTFGFSPLATALLHEFGTWQGANLLLAGLILNCAIFGALMRPLVYPKTSGEKPLLQRMAEEKRLQMERGSIGGSYFVVQLPDGTMEKRLKAPLNIDPGVHSSLNLEALARVPTVPNMHGVPTVPTLPTITEAKVVDTTDKKKNENGNVIVNPVQVQISRNVSSPAFSAQAPGLPKNGSVPFFDRQRKHSTGEKGAERFKPSLAAIKATSKTSVGSHRPTPDGDTESNMYNSKLSVSGPREAPRMVRPMSRKDIFYSGSVLNLPQYQSQKSLQGYRNSVLSLPQSRQTGDLERQEQYDLCPCLHLPNSFKSALSSMLDVSLLKDPAFMLIGVSNIFGMAGLYVPFVYLTDAAKVNGIEPSQASFLLSIIGITNTFGRIICGWVADFPWMDSLMLNNVCLVIATVSVGVTPLCTTYSAYVVIAITFGIAISGYISLTSIILVDLLGLEKLTNAFGLLILFRGAAAMVGSPLAGAVFDATGTYGPSFYMASVFFLISTLTSFAAPLFRRKQEEIPQPLDVLTPIDEDLEEGDEEDPDDTPITMGAQLASRTPAITRTAASPSDPPSPTNNELQQRESVL
Summary
Uniprot
H9JPX1
A0A2H1V3Z5
A0A3S2LWE8
A0A194PSF3
A0A212FH79
A0A1E1WTA4
+ More
A0A194RIH5 A0A1E1VZQ4 A0A0K8TKH4 U5ETH5 A0A1L8DEB7 A0A182PE33 A0A1L8DEF7 A0A1L8DEK1 A0A3F2Z011 A0A182N5C0 A0A1L8DEP1 A0A182RIV8 A0A2M4CTS0 A0A182VRH1 A0A182VCP8 A0A182JGN4 A0A182FSL6 A0A182LCG6 A0A182XVZ8 A0A182Q9L7 A0A2M4CUA9 A0A182HJ88 A0A2M4BDZ0 A0A2M4CRU6 Q7QBE4 A0A2M4AFB1 F5HL15 A0A0P6IZT9 A0A182XE34 A0A2M4CRY8 W8AV54 A0A034V8T1 A0A2M4CRS1 A0A0A1XFG7 A0A0K8UB75 A0A0K8UT30 A0A1Q3FML3 A0A1Q3FMN2 A0A1Q3FMJ4 B0W6P2 A0A182LYT1 A0A182GZ04 A0A1Q3FMD6 A0A1Q3FM91 A0A182K398 A0A1S4EVS2 Q17P96 A0A3L8DUV7 A0A195BFX9 E9J9H8 F4WHW1 A0A195FL03 A0A195C9U1 A0A151IUR6 A0A2A3EKB1 A0A158NBY4 A0A084WSJ5 A0A1B0FB13 A0A1B0AIH7 A0A1B0B0D3 A0A0L0C4T4 A0A1A9XT47 A0A154P4Y2 U4UGB1 A0A1A9VAM8 A0A1A9W537 A0A1Y1LMX7 A0A023F419 A0A0L7R1U4 A0A088A5U8 A0A1B6C6S6 A0A224XIS1 A0A0P4VSG9 T1PDQ5 B0WYT3 A0A139WNL4 K7IRA0 A0A0C9Q106 A0A1B6DUM1 B4ML46 A0A0C9RSN1 T1IFT8 A0A1I8NYR6 B4HBN2 Q2LZU9 A0A026VY47 D6WBF2 E1ZY63 A0A067RDK0 A0A3B0KF50 B3M5E4 E2BUR4 A0A1W4XG00 A0A1J1HE30
A0A194RIH5 A0A1E1VZQ4 A0A0K8TKH4 U5ETH5 A0A1L8DEB7 A0A182PE33 A0A1L8DEF7 A0A1L8DEK1 A0A3F2Z011 A0A182N5C0 A0A1L8DEP1 A0A182RIV8 A0A2M4CTS0 A0A182VRH1 A0A182VCP8 A0A182JGN4 A0A182FSL6 A0A182LCG6 A0A182XVZ8 A0A182Q9L7 A0A2M4CUA9 A0A182HJ88 A0A2M4BDZ0 A0A2M4CRU6 Q7QBE4 A0A2M4AFB1 F5HL15 A0A0P6IZT9 A0A182XE34 A0A2M4CRY8 W8AV54 A0A034V8T1 A0A2M4CRS1 A0A0A1XFG7 A0A0K8UB75 A0A0K8UT30 A0A1Q3FML3 A0A1Q3FMN2 A0A1Q3FMJ4 B0W6P2 A0A182LYT1 A0A182GZ04 A0A1Q3FMD6 A0A1Q3FM91 A0A182K398 A0A1S4EVS2 Q17P96 A0A3L8DUV7 A0A195BFX9 E9J9H8 F4WHW1 A0A195FL03 A0A195C9U1 A0A151IUR6 A0A2A3EKB1 A0A158NBY4 A0A084WSJ5 A0A1B0FB13 A0A1B0AIH7 A0A1B0B0D3 A0A0L0C4T4 A0A1A9XT47 A0A154P4Y2 U4UGB1 A0A1A9VAM8 A0A1A9W537 A0A1Y1LMX7 A0A023F419 A0A0L7R1U4 A0A088A5U8 A0A1B6C6S6 A0A224XIS1 A0A0P4VSG9 T1PDQ5 B0WYT3 A0A139WNL4 K7IRA0 A0A0C9Q106 A0A1B6DUM1 B4ML46 A0A0C9RSN1 T1IFT8 A0A1I8NYR6 B4HBN2 Q2LZU9 A0A026VY47 D6WBF2 E1ZY63 A0A067RDK0 A0A3B0KF50 B3M5E4 E2BUR4 A0A1W4XG00 A0A1J1HE30
Pubmed
EMBL
BABH01015445
BABH01015446
ODYU01000573
SOQ35565.1
RSAL01000149
RVE45813.1
+ More
KQ459601 KPI94055.1 AGBW02008541 OWR53063.1 GDQN01000887 JAT90167.1 KQ460205 KPJ17130.1 GDQN01010838 JAT80216.1 GDAI01002804 JAI14799.1 GANO01004371 JAB55500.1 GFDF01009275 JAV04809.1 GFDF01009276 JAV04808.1 GFDF01009192 JAV04892.1 GFDF01009152 JAV04932.1 GGFL01004491 MBW68669.1 AXCN02000148 GGFL01004754 MBW68932.1 APCN01002131 GGFJ01002125 MBW51266.1 GGFL01003855 MBW68033.1 AAAB01008879 EAA08484.4 GGFK01006144 MBW39465.1 EGK96976.1 GDUN01000592 JAN95327.1 GGFL01003857 MBW68035.1 GAMC01016429 GAMC01016428 JAB90127.1 GAKP01020420 JAC38532.1 GGFL01003856 MBW68034.1 GBXI01004611 JAD09681.1 GDHF01028714 GDHF01024954 JAI23600.1 JAI27360.1 GDHF01025720 GDHF01022799 JAI26594.1 JAI29515.1 GFDL01006260 JAV28785.1 GFDL01006176 JAV28869.1 GFDL01006280 JAV28765.1 DS231849 EDS36873.1 AXCM01002065 JXUM01021819 JXUM01021820 KQ560612 KXJ81624.1 GFDL01006382 JAV28663.1 GFDL01006439 JAV28606.1 CH477193 EAT48536.1 QOIP01000004 RLU23538.1 KQ976500 KYM83087.1 GL769313 EFZ10528.1 GL888170 EGI66190.1 KQ981491 KYN41026.1 KQ978068 KYM97480.1 KQ980949 KYN11211.1 KZ288229 PBC31692.1 ADTU01011486 ADTU01011487 ADTU01011488 ADTU01011489 ATLV01026582 ATLV01026583 ATLV01026584 ATLV01026585 ATLV01026586 ATLV01026587 ATLV01026588 ATLV01026589 ATLV01026590 KE525415 KFB53189.1 CCAG010023620 JXJN01006745 JRES01000919 KNC27251.1 KQ434809 KZC06190.1 KB632184 ERL89646.1 GEZM01054376 JAV73710.1 GBBI01002492 JAC16220.1 KQ414667 KOC64808.1 GEDC01028095 JAS09203.1 GFTR01008086 JAW08340.1 GDKW01001064 JAI55531.1 KA646829 AFP61458.1 DS232198 EDS37193.1 KQ971312 KYB29401.1 GBYB01007593 JAG77360.1 GEDC01022746 GEDC01007924 JAS14552.1 JAS29374.1 CH963847 EDW73104.1 GBYB01010486 JAG80253.1 ACPB03001166 CH479261 EDW39436.1 CH379069 EAL31188.2 KK107785 EZA47789.1 EEZ98726.1 GL435171 EFN73912.1 KK852771 KDR16894.1 OUUW01000009 SPP84939.1 CH902618 EDV39554.1 GL450743 EFN80569.1 CVRI01000001 CRK86207.1
KQ459601 KPI94055.1 AGBW02008541 OWR53063.1 GDQN01000887 JAT90167.1 KQ460205 KPJ17130.1 GDQN01010838 JAT80216.1 GDAI01002804 JAI14799.1 GANO01004371 JAB55500.1 GFDF01009275 JAV04809.1 GFDF01009276 JAV04808.1 GFDF01009192 JAV04892.1 GFDF01009152 JAV04932.1 GGFL01004491 MBW68669.1 AXCN02000148 GGFL01004754 MBW68932.1 APCN01002131 GGFJ01002125 MBW51266.1 GGFL01003855 MBW68033.1 AAAB01008879 EAA08484.4 GGFK01006144 MBW39465.1 EGK96976.1 GDUN01000592 JAN95327.1 GGFL01003857 MBW68035.1 GAMC01016429 GAMC01016428 JAB90127.1 GAKP01020420 JAC38532.1 GGFL01003856 MBW68034.1 GBXI01004611 JAD09681.1 GDHF01028714 GDHF01024954 JAI23600.1 JAI27360.1 GDHF01025720 GDHF01022799 JAI26594.1 JAI29515.1 GFDL01006260 JAV28785.1 GFDL01006176 JAV28869.1 GFDL01006280 JAV28765.1 DS231849 EDS36873.1 AXCM01002065 JXUM01021819 JXUM01021820 KQ560612 KXJ81624.1 GFDL01006382 JAV28663.1 GFDL01006439 JAV28606.1 CH477193 EAT48536.1 QOIP01000004 RLU23538.1 KQ976500 KYM83087.1 GL769313 EFZ10528.1 GL888170 EGI66190.1 KQ981491 KYN41026.1 KQ978068 KYM97480.1 KQ980949 KYN11211.1 KZ288229 PBC31692.1 ADTU01011486 ADTU01011487 ADTU01011488 ADTU01011489 ATLV01026582 ATLV01026583 ATLV01026584 ATLV01026585 ATLV01026586 ATLV01026587 ATLV01026588 ATLV01026589 ATLV01026590 KE525415 KFB53189.1 CCAG010023620 JXJN01006745 JRES01000919 KNC27251.1 KQ434809 KZC06190.1 KB632184 ERL89646.1 GEZM01054376 JAV73710.1 GBBI01002492 JAC16220.1 KQ414667 KOC64808.1 GEDC01028095 JAS09203.1 GFTR01008086 JAW08340.1 GDKW01001064 JAI55531.1 KA646829 AFP61458.1 DS232198 EDS37193.1 KQ971312 KYB29401.1 GBYB01007593 JAG77360.1 GEDC01022746 GEDC01007924 JAS14552.1 JAS29374.1 CH963847 EDW73104.1 GBYB01010486 JAG80253.1 ACPB03001166 CH479261 EDW39436.1 CH379069 EAL31188.2 KK107785 EZA47789.1 EEZ98726.1 GL435171 EFN73912.1 KK852771 KDR16894.1 OUUW01000009 SPP84939.1 CH902618 EDV39554.1 GL450743 EFN80569.1 CVRI01000001 CRK86207.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000075885
+ More
UP000075903 UP000075884 UP000075900 UP000075920 UP000075880 UP000069272 UP000075882 UP000076408 UP000075886 UP000075840 UP000007062 UP000076407 UP000002320 UP000075883 UP000069940 UP000249989 UP000075881 UP000008820 UP000279307 UP000078540 UP000007755 UP000078541 UP000078542 UP000078492 UP000242457 UP000005205 UP000030765 UP000092444 UP000092445 UP000092460 UP000037069 UP000092443 UP000076502 UP000030742 UP000078200 UP000091820 UP000053825 UP000005203 UP000095301 UP000007266 UP000002358 UP000007798 UP000015103 UP000095300 UP000008744 UP000001819 UP000053097 UP000000311 UP000027135 UP000268350 UP000007801 UP000008237 UP000192223 UP000183832
UP000075903 UP000075884 UP000075900 UP000075920 UP000075880 UP000069272 UP000075882 UP000076408 UP000075886 UP000075840 UP000007062 UP000076407 UP000002320 UP000075883 UP000069940 UP000249989 UP000075881 UP000008820 UP000279307 UP000078540 UP000007755 UP000078541 UP000078542 UP000078492 UP000242457 UP000005205 UP000030765 UP000092444 UP000092445 UP000092460 UP000037069 UP000092443 UP000076502 UP000030742 UP000078200 UP000091820 UP000053825 UP000005203 UP000095301 UP000007266 UP000002358 UP000007798 UP000015103 UP000095300 UP000008744 UP000001819 UP000053097 UP000000311 UP000027135 UP000268350 UP000007801 UP000008237 UP000192223 UP000183832
PRIDE
Pfam
PF07690 MFS_1
CDD
ProteinModelPortal
H9JPX1
A0A2H1V3Z5
A0A3S2LWE8
A0A194PSF3
A0A212FH79
A0A1E1WTA4
+ More
A0A194RIH5 A0A1E1VZQ4 A0A0K8TKH4 U5ETH5 A0A1L8DEB7 A0A182PE33 A0A1L8DEF7 A0A1L8DEK1 A0A3F2Z011 A0A182N5C0 A0A1L8DEP1 A0A182RIV8 A0A2M4CTS0 A0A182VRH1 A0A182VCP8 A0A182JGN4 A0A182FSL6 A0A182LCG6 A0A182XVZ8 A0A182Q9L7 A0A2M4CUA9 A0A182HJ88 A0A2M4BDZ0 A0A2M4CRU6 Q7QBE4 A0A2M4AFB1 F5HL15 A0A0P6IZT9 A0A182XE34 A0A2M4CRY8 W8AV54 A0A034V8T1 A0A2M4CRS1 A0A0A1XFG7 A0A0K8UB75 A0A0K8UT30 A0A1Q3FML3 A0A1Q3FMN2 A0A1Q3FMJ4 B0W6P2 A0A182LYT1 A0A182GZ04 A0A1Q3FMD6 A0A1Q3FM91 A0A182K398 A0A1S4EVS2 Q17P96 A0A3L8DUV7 A0A195BFX9 E9J9H8 F4WHW1 A0A195FL03 A0A195C9U1 A0A151IUR6 A0A2A3EKB1 A0A158NBY4 A0A084WSJ5 A0A1B0FB13 A0A1B0AIH7 A0A1B0B0D3 A0A0L0C4T4 A0A1A9XT47 A0A154P4Y2 U4UGB1 A0A1A9VAM8 A0A1A9W537 A0A1Y1LMX7 A0A023F419 A0A0L7R1U4 A0A088A5U8 A0A1B6C6S6 A0A224XIS1 A0A0P4VSG9 T1PDQ5 B0WYT3 A0A139WNL4 K7IRA0 A0A0C9Q106 A0A1B6DUM1 B4ML46 A0A0C9RSN1 T1IFT8 A0A1I8NYR6 B4HBN2 Q2LZU9 A0A026VY47 D6WBF2 E1ZY63 A0A067RDK0 A0A3B0KF50 B3M5E4 E2BUR4 A0A1W4XG00 A0A1J1HE30
A0A194RIH5 A0A1E1VZQ4 A0A0K8TKH4 U5ETH5 A0A1L8DEB7 A0A182PE33 A0A1L8DEF7 A0A1L8DEK1 A0A3F2Z011 A0A182N5C0 A0A1L8DEP1 A0A182RIV8 A0A2M4CTS0 A0A182VRH1 A0A182VCP8 A0A182JGN4 A0A182FSL6 A0A182LCG6 A0A182XVZ8 A0A182Q9L7 A0A2M4CUA9 A0A182HJ88 A0A2M4BDZ0 A0A2M4CRU6 Q7QBE4 A0A2M4AFB1 F5HL15 A0A0P6IZT9 A0A182XE34 A0A2M4CRY8 W8AV54 A0A034V8T1 A0A2M4CRS1 A0A0A1XFG7 A0A0K8UB75 A0A0K8UT30 A0A1Q3FML3 A0A1Q3FMN2 A0A1Q3FMJ4 B0W6P2 A0A182LYT1 A0A182GZ04 A0A1Q3FMD6 A0A1Q3FM91 A0A182K398 A0A1S4EVS2 Q17P96 A0A3L8DUV7 A0A195BFX9 E9J9H8 F4WHW1 A0A195FL03 A0A195C9U1 A0A151IUR6 A0A2A3EKB1 A0A158NBY4 A0A084WSJ5 A0A1B0FB13 A0A1B0AIH7 A0A1B0B0D3 A0A0L0C4T4 A0A1A9XT47 A0A154P4Y2 U4UGB1 A0A1A9VAM8 A0A1A9W537 A0A1Y1LMX7 A0A023F419 A0A0L7R1U4 A0A088A5U8 A0A1B6C6S6 A0A224XIS1 A0A0P4VSG9 T1PDQ5 B0WYT3 A0A139WNL4 K7IRA0 A0A0C9Q106 A0A1B6DUM1 B4ML46 A0A0C9RSN1 T1IFT8 A0A1I8NYR6 B4HBN2 Q2LZU9 A0A026VY47 D6WBF2 E1ZY63 A0A067RDK0 A0A3B0KF50 B3M5E4 E2BUR4 A0A1W4XG00 A0A1J1HE30
Ontologies
GO
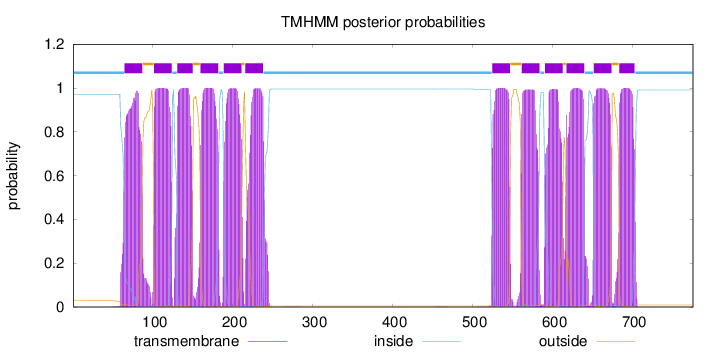
Topology
Length:
776
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
264.84309
Exp number, first 60 AAs:
0.17448
Total prob of N-in:
0.97152
inside
1 - 64
TMhelix
65 - 87
outside
88 - 101
TMhelix
102 - 124
inside
125 - 130
TMhelix
131 - 150
outside
151 - 159
TMhelix
160 - 182
inside
183 - 188
TMhelix
189 - 211
outside
212 - 215
TMhelix
216 - 238
inside
239 - 524
TMhelix
525 - 547
outside
548 - 561
TMhelix
562 - 584
inside
585 - 590
TMhelix
591 - 613
outside
614 - 617
TMhelix
618 - 640
inside
641 - 651
TMhelix
652 - 674
outside
675 - 683
TMhelix
684 - 703
inside
704 - 776
Population Genetic Test Statistics
Pi
212.42617
Theta
168.736631
Tajima's D
0.953462
CLR
0.091032
CSRT
0.646467676616169
Interpretation
Uncertain
