Gene
KWMTBOMO13853 Validated by peptides from experiments
Annotation
Agrin_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 2.66
Sequence
CDS
ATGAGACAACACGTCTTTTTCAAAGCAACATTCGCCATACTGTTCATTCAAGTCACGGACTGCTATGAATGGCACAACAGGAAAAACCAAGATCACGGCGATTGGTTTCACCACGGATCGCGACAATGGTCAACAAGGAGGAAAGATTCCCACGATTGGGGTTCGAGCGAGTCTGCAAGCAGCGAAGAATCAGACTATGGTGCTACACATTATGGGGGAGGAAATCACAGGAACTGGCAACCTTGGGGACACAATTTTCACTATCCATCTGGCCAGCATGAACATCGGTATCGGCCCAATTTTCATGCATATGACTTGAACCTCAGACCGGAATTCAATGGGAATAGATTTAACAGACAAACGACCGCAACCGCCGGGTCTCCGGACATCGAAGCCTGCATCCGTAGTTGTCCTGTTACATCAGAATATAATCCTGTTTGTGGTACCAATAATGAGACCTACAATAATTTAGGAAGATTAATGTGTGCTCAGCAATGCGGAGTTGTTGTTGAAATACAACGGAAAAGTACGTGCTCGGCAGCAAACGTCCCTACTGAATCATCGACAAGTTTCGACAGAGGAGATGGTCTTCTACAGACTACTGATGTCCCTGCCAATATCCAGCAATGCATTTCTTTATGTCCTCAATCAGAGGAAACGAGGCCTGTTTGTGCTTCAAACGGAGTCACATATAGAAATCCATTGGCAATCCTTTGTGCTCAGATGTGTGGACTTGACATCAGAGCCACGAAGTTGTCAGCATGCACAATCGATAATCGAAAGCCCGTGGTCCAACAGCCACTGTCGCTCAGTACCCTGACATGTATTAAATCCTGTCCCACCACCACTGAGAACAAACCAGTATGTGGAACCAATGGCGTGACATATCAGAACCCAAGCTATATACAATGCATTCGTATTTGTGGAGTCAATGTGGAGATACATAGAAGAGCTCCGTGCCCTTCATCAGCCACTAATGACGTTCAAACTACTGCAATGCTGTCACCGGACGAGTTACTTTTGACTCCAGCAGTCCAATTTTGTATAAAGTCTTGTCCTGCGACATCTGAATTCAACCCTGTTTGTGGGACGAACTTAGTGACGTACATGAACCCCAGTAGATTGCAATGTGCTAGAACATGTGGAGTCAAGGTGGAGATAATGCAAAGACGAAAATGCCCATCGATCGTATTACCTAATAGGGAGCCGAATGACGACGGAAACAATAATATTCCCTTGCCTAATGTACCTTCTAGTACGGTACCTCCATGGAAGCCGACGATAGGAGAATGCTTGGCCGCTTGTCCGCAATCACAGATATATGAACCCATTTGTGGGACCAATAACTTTACTTTTAACAATGCGGACCATTTGCTTTGTGCACAGTTATGTGGAATTGATGTGCAAATTCTACATCAATCCGAGTGTACCAGTGGCGAGGTTACTTCAGAGACAATTCCGCCTCTGACGACCGTGTCGCCTGATAATCAACCAAACAATAACGAAGAAACTACAGAGAAGCTTGTAGTACCTATAAACGAATCTTCAAGTTCTCCACCTTTCACTATACCCTCTGATATAATTAACAATATTTTCACTGAGCCCACTACGTTAGATCCTACAGAAGATATTGATCAAAGATTTGGTGACAAATAA
Protein
MRQHVFFKATFAILFIQVTDCYEWHNRKNQDHGDWFHHGSRQWSTRRKDSHDWGSSESASSEESDYGATHYGGGNHRNWQPWGHNFHYPSGQHEHRYRPNFHAYDLNLRPEFNGNRFNRQTTATAGSPDIEACIRSCPVTSEYNPVCGTNNETYNNLGRLMCAQQCGVVVEIQRKSTCSAANVPTESSTSFDRGDGLLQTTDVPANIQQCISLCPQSEETRPVCASNGVTYRNPLAILCAQMCGLDIRATKLSACTIDNRKPVVQQPLSLSTLTCIKSCPTTTENKPVCGTNGVTYQNPSYIQCIRICGVNVEIHRRAPCPSSATNDVQTTAMLSPDELLLTPAVQFCIKSCPATSEFNPVCGTNLVTYMNPSRLQCARTCGVKVEIMQRRKCPSIVLPNREPNDDGNNNIPLPNVPSSTVPPWKPTIGECLAACPQSQIYEPICGTNNFTFNNADHLLCAQLCGIDVQILHQSECTSGEVTSETIPPLTTVSPDNQPNNNEETTEKLVVPINESSSSPPFTIPSDIINNIFTEPTTLDPTEDIDQRFGDK
Summary
Uniprot
EMBL
BABH01015445
JTDY01006172
KOB66223.1
JTDY01003433
KOB69562.1
DQ236243
+ More
ABB58758.1 EU183309 ACD88987.1 DF237678 GAQ91113.1 AHZU02001279 KFG34074.1 AFHV02003173 PUA84448.1 AEYH02002314 KFG40610.1 AEYI02001212 KFG40175.1 AFYV02000360 KFG65263.1 AEYJ02000356 KFH10805.1 AHZP02000549 KYK70386.1 AAQM03000090 EPR62761.1 LN714497 AAYL02000145 CEL74369.1 ESS32012.1
ABB58758.1 EU183309 ACD88987.1 DF237678 GAQ91113.1 AHZU02001279 KFG34074.1 AFHV02003173 PUA84448.1 AEYH02002314 KFG40610.1 AEYI02001212 KFG40175.1 AFYV02000360 KFG65263.1 AEYJ02000356 KFH10805.1 AHZP02000549 KYK70386.1 AAQM03000090 EPR62761.1 LN714497 AAYL02000145 CEL74369.1 ESS32012.1
Proteomes
PRIDE
SUPFAM
SSF100895
SSF100895
ProteinModelPortal
PDB
2P6A
E-value=0.00149305,
Score=100
Ontologies
GO
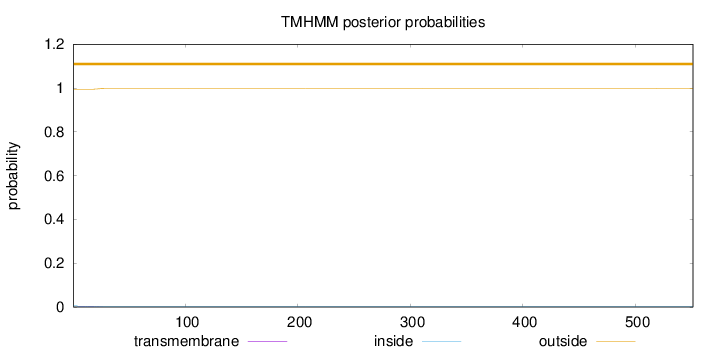
Topology
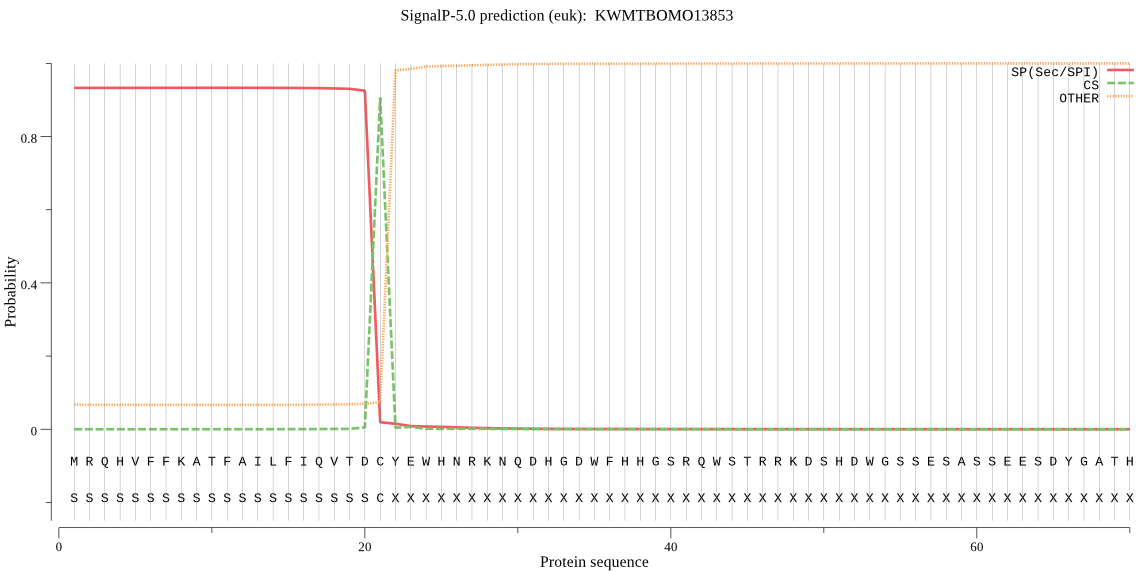
SignalP
Position: 1 - 21,
Likelihood: 0.933038
Length:
551
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0544500000000001
Exp number, first 60 AAs:
0.05425
Total prob of N-in:
0.00537
outside
1 - 551
Population Genetic Test Statistics
Pi
196.072941
Theta
160.070939
Tajima's D
0.798279
CLR
0.104524
CSRT
0.602369881505925
Interpretation
Uncertain
