Gene
KWMTBOMO13852 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011574
Annotation
protease_inhibitor_1_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.595
Sequence
CDS
ATGGATAAATTAGTAGTATTTTTCCTTTTCGCAATCATAACTAACGTGTTATGTATGAGCGTAAGGAACAAGCGTCAGTCGAATGATGATGATGACGTTCTCGATGACCGCTATGGCTGGGAGCTTACCACCCGGCCTCCAAGGCAGTTCCCTGGGCAAGGATTTTTCCCCGGTCTATTCCCCGGCCAGGGTCAGTTCCCAGGACAACAGCAACGTTTAACTACGACTCGGGCTCCCAACAATCTGGGCACCACCACAATGTCGCCTGCAATTCAACAATGCATTCGTAGCTGCCCAGTAACCGCTGAGTACAATCCAGTTTGTGGCACTGATAATATAACTTACAATAACCCTGGAAGGTTGACGTGTGCTCAGGCGTGTGGAATCAATGTCAGCGTTCTCCGATCCCTGCCTTGCCCCACTGCTACACAAGCTCCTACCAGCTAA
Protein
MDKLVVFFLFAIITNVLCMSVRNKRQSNDDDDVLDDRYGWELTTRPPRQFPGQGFFPGLFPGQGQFPGQQQRLTTTRAPNNLGTTTMSPAIQQCIRSCPVTAEYNPVCGTDNITYNNPGRLTCAQACGINVSVLRSLPCPTATQAPTS
Summary
Uniprot
EMBL
Proteomes
SUPFAM
SSF100895
SSF100895
ProteinModelPortal
PDB
1TBR
E-value=0.0387441,
Score=80
Ontologies
GO
Topology
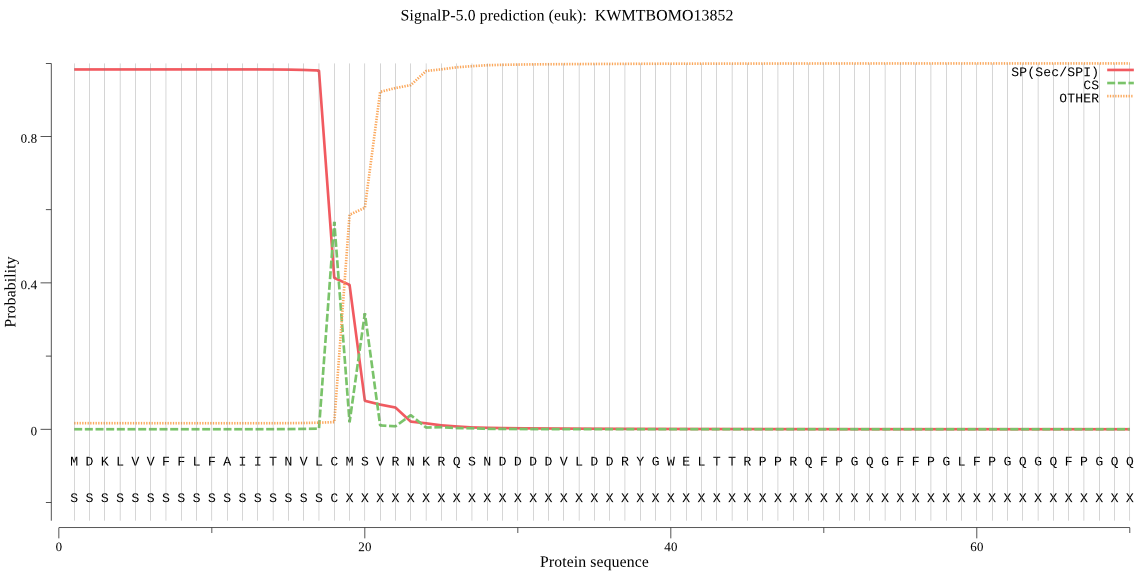
SignalP
Position: 1 - 18,
Likelihood: 0.983289
Length:
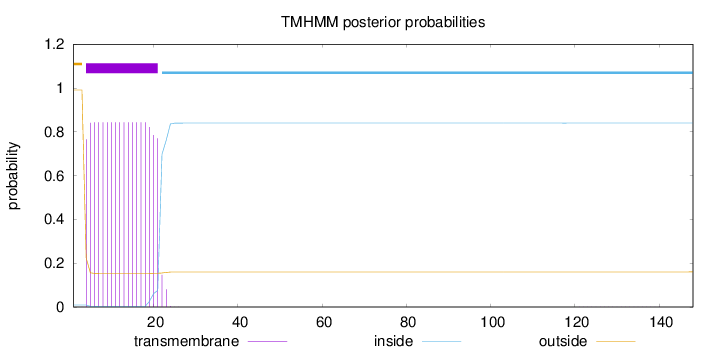
148
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.19191
Exp number, first 60 AAs:
15.18655
Total prob of N-in:
0.00923
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 21
inside
22 - 148
Population Genetic Test Statistics
Pi
167.523686
Theta
152.554281
Tajima's D
0.948049
CLR
0.821056
CSRT
0.64376781160942
Interpretation
Uncertain
