Gene
KWMTBOMO13850 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011573
Annotation
kazal-type_proteinase_inhibitor_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.672
Sequence
CDS
ATGAAGACCATTATTTTCTGTGCTTTCGCTGTGTTGTTCGTGGCGGTGTCGTGCCGTCCCGACAAGCTGGACCTGAAGCAGGTGAAGGCTGATGCTGAGCGCAAGAAGGCCTGCATGCATGACTGCACCAAAGCTAATCTTGACCCTATCTGCGCTGGAAAGACAGGAGAGAAACCCAAGTCATTCGGAAACGAGTGTGTCATGAACAACTACAATTGCGAACACAAAGATACGTTGCGCAAGATCAGCCAAGGCCAGTGTCCTGGATCAGATGGCATCCGTCTCTCTTGA
Protein
MKTIIFCAFAVLFVAVSCRPDKLDLKQVKADAERKKACMHDCTKANLDPICAGKTGEKPKSFGNECVMNNYNCEHKDTLRKISQGQCPGSDGIRLS
Summary
Uniprot
Q2F5Q6
A0A2H1VFQ4
S4P713
A0A2A4JIZ0
Q9U512
A0A194PL44
+ More
A0A194RR44 I4DJ04 A0A1V0CMG0 Q0Q007 A0A059VA86 A0A212EH58 A0A3S2NR65 Q5MGF1 U4U954 A0A023EEC1 B0W0R4 S4TSP0 A0A1L8DPD8 A0A1L8DPG5 A0A139WMV7 Q17PZ7 A0A1B0EUB7 T1E3E4 A0A1B6IC76 A0A1B6MTB9 A0A1B6LVK6 V9IFN0 A0A195F2V2 A0A1I8MTJ0 Q0PXV7 A0A1I8NPP4 A0A0L0BZW5 A0A0M9A936 A0A336MEN6 B4N9G7 B4K9D5 A0A1B0C5K0 A0A0M3QY17 A0A1W4VRI7 B3LXY2 B4LXK0 A0A182H3D5 A0A3B0KH47 A0A182FLX9 T1EB65 B4K7L8 A0A2M3YWJ7 A0A2M3YWI2 B6DE58
A0A194RR44 I4DJ04 A0A1V0CMG0 Q0Q007 A0A059VA86 A0A212EH58 A0A3S2NR65 Q5MGF1 U4U954 A0A023EEC1 B0W0R4 S4TSP0 A0A1L8DPD8 A0A1L8DPG5 A0A139WMV7 Q17PZ7 A0A1B0EUB7 T1E3E4 A0A1B6IC76 A0A1B6MTB9 A0A1B6LVK6 V9IFN0 A0A195F2V2 A0A1I8MTJ0 Q0PXV7 A0A1I8NPP4 A0A0L0BZW5 A0A0M9A936 A0A336MEN6 B4N9G7 B4K9D5 A0A1B0C5K0 A0A0M3QY17 A0A1W4VRI7 B3LXY2 B4LXK0 A0A182H3D5 A0A3B0KH47 A0A182FLX9 T1EB65 B4K7L8 A0A2M3YWJ7 A0A2M3YWI2 B6DE58
Pubmed
EMBL
BABH01015442
DQ311367
ABD36311.1
ODYU01002301
SOQ39616.1
GAIX01006616
+ More
JAA85944.1 NWSH01001221 PCG72065.1 AF117576 AAF16698.1 KQ459601 KPI94052.1 KQ459833 KPJ19780.1 AK401272 BAM17894.1 KY673262 ARA71543.1 DQ666524 ABG72727.1 KJ546374 AHZ90949.1 AGBW02014956 OWR40814.1 RSAL01000006 RVE54230.1 AY829835 AAV91449.1 KB631815 ERL86470.1 GAPW01006634 JAC06964.1 DS231818 EDS41366.1 AJVK01033734 JX171681 AGF86395.1 GFDF01005840 JAV08244.1 GFDF01005839 JAV08245.1 KQ971312 KYB29223.1 CH477188 EAT48797.1 AJWK01014206 GALA01000430 JAA94422.1 GECU01023204 JAS84502.1 GEBQ01000797 JAT39180.1 GEBQ01012283 JAT27694.1 JR040779 JR040780 AEY59290.1 AEY59291.1 KQ981855 KYN34905.1 DQ673429 ABG82002.1 JRES01001097 KNC25558.1 KQ435710 KOX79717.1 UFQT01001076 SSX28765.1 CH964232 EDW81643.1 CH933806 EDW15567.1 JXJN01026042 JXJN01026043 CP012526 ALC46893.1 CH902617 EDV42838.1 CH940650 EDW66784.1 JXUM01107201 KQ565122 KXJ71313.1 OUUW01000008 SPP84421.1 GAMD01000238 JAB01353.1 EDW15362.2 GGFF01000164 MBW20631.1 GGFF01000139 MBW20606.1 EU934427 ACI30205.1
JAA85944.1 NWSH01001221 PCG72065.1 AF117576 AAF16698.1 KQ459601 KPI94052.1 KQ459833 KPJ19780.1 AK401272 BAM17894.1 KY673262 ARA71543.1 DQ666524 ABG72727.1 KJ546374 AHZ90949.1 AGBW02014956 OWR40814.1 RSAL01000006 RVE54230.1 AY829835 AAV91449.1 KB631815 ERL86470.1 GAPW01006634 JAC06964.1 DS231818 EDS41366.1 AJVK01033734 JX171681 AGF86395.1 GFDF01005840 JAV08244.1 GFDF01005839 JAV08245.1 KQ971312 KYB29223.1 CH477188 EAT48797.1 AJWK01014206 GALA01000430 JAA94422.1 GECU01023204 JAS84502.1 GEBQ01000797 JAT39180.1 GEBQ01012283 JAT27694.1 JR040779 JR040780 AEY59290.1 AEY59291.1 KQ981855 KYN34905.1 DQ673429 ABG82002.1 JRES01001097 KNC25558.1 KQ435710 KOX79717.1 UFQT01001076 SSX28765.1 CH964232 EDW81643.1 CH933806 EDW15567.1 JXJN01026042 JXJN01026043 CP012526 ALC46893.1 CH902617 EDV42838.1 CH940650 EDW66784.1 JXUM01107201 KQ565122 KXJ71313.1 OUUW01000008 SPP84421.1 GAMD01000238 JAB01353.1 EDW15362.2 GGFF01000164 MBW20631.1 GGFF01000139 MBW20606.1 EU934427 ACI30205.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000030742 UP000002320 UP000092462 UP000007266 UP000008820 UP000092461 UP000078541 UP000095301 UP000079169 UP000095300 UP000037069 UP000053105 UP000007798 UP000009192 UP000092460 UP000092553 UP000192221 UP000007801 UP000008792 UP000069940 UP000249989 UP000268350 UP000069272
UP000030742 UP000002320 UP000092462 UP000007266 UP000008820 UP000092461 UP000078541 UP000095301 UP000079169 UP000095300 UP000037069 UP000053105 UP000007798 UP000009192 UP000092460 UP000092553 UP000192221 UP000007801 UP000008792 UP000069940 UP000249989 UP000268350 UP000069272
PRIDE
SUPFAM
SSF100895
SSF100895
ProteinModelPortal
Q2F5Q6
A0A2H1VFQ4
S4P713
A0A2A4JIZ0
Q9U512
A0A194PL44
+ More
A0A194RR44 I4DJ04 A0A1V0CMG0 Q0Q007 A0A059VA86 A0A212EH58 A0A3S2NR65 Q5MGF1 U4U954 A0A023EEC1 B0W0R4 S4TSP0 A0A1L8DPD8 A0A1L8DPG5 A0A139WMV7 Q17PZ7 A0A1B0EUB7 T1E3E4 A0A1B6IC76 A0A1B6MTB9 A0A1B6LVK6 V9IFN0 A0A195F2V2 A0A1I8MTJ0 Q0PXV7 A0A1I8NPP4 A0A0L0BZW5 A0A0M9A936 A0A336MEN6 B4N9G7 B4K9D5 A0A1B0C5K0 A0A0M3QY17 A0A1W4VRI7 B3LXY2 B4LXK0 A0A182H3D5 A0A3B0KH47 A0A182FLX9 T1EB65 B4K7L8 A0A2M3YWJ7 A0A2M3YWI2 B6DE58
A0A194RR44 I4DJ04 A0A1V0CMG0 Q0Q007 A0A059VA86 A0A212EH58 A0A3S2NR65 Q5MGF1 U4U954 A0A023EEC1 B0W0R4 S4TSP0 A0A1L8DPD8 A0A1L8DPG5 A0A139WMV7 Q17PZ7 A0A1B0EUB7 T1E3E4 A0A1B6IC76 A0A1B6MTB9 A0A1B6LVK6 V9IFN0 A0A195F2V2 A0A1I8MTJ0 Q0PXV7 A0A1I8NPP4 A0A0L0BZW5 A0A0M9A936 A0A336MEN6 B4N9G7 B4K9D5 A0A1B0C5K0 A0A0M3QY17 A0A1W4VRI7 B3LXY2 B4LXK0 A0A182H3D5 A0A3B0KH47 A0A182FLX9 T1EB65 B4K7L8 A0A2M3YWJ7 A0A2M3YWI2 B6DE58
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.998091
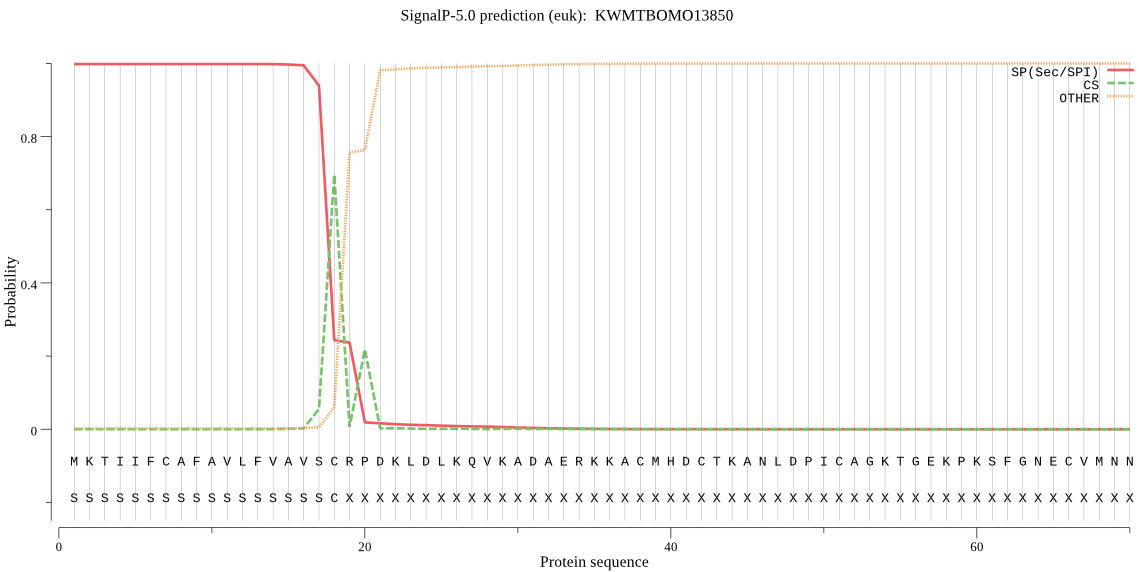
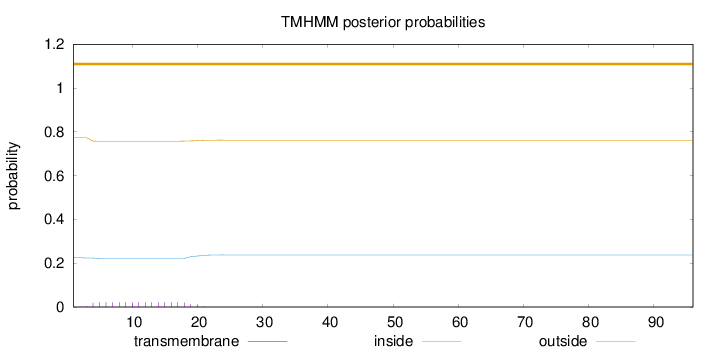
Length:
96
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.34861
Exp number, first 60 AAs:
0.34861
Total prob of N-in:
0.22705
outside
1 - 96
Population Genetic Test Statistics
Pi
52.174609
Theta
162.879729
Tajima's D
-1.599063
CLR
2290.393276
CSRT
0.0487475626218689
Interpretation
Possibly Positive selection
