Gene
KWMTBOMO13848
Pre Gene Modal
BGIBMGA011569
Annotation
PREDICTED:_lysine-specific_demethylase_3B_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.418 Mitochondrial Reliability : 1.53
Sequence
CDS
ATGGCCTATAAATATCGTGAAGATCTAGTTGGCAAACGATTTCTCGCAGTGAGTGGTTTAACTAAAATTAATGTGAATAGAGTTTCCGAATGGGGTTGGAAAGCTGGTGTTATCAGAGCAGCATCACTCAGAGATAACAAGAATAAGGAACTCCAGGTTTTAGTGGAATACGATGGTGTAGAATGGCAACGACGCGAGTGGGTAGCTGTGTATTCGCGACGAACTTTTCGCGTGTTTTTAGTAGAAAGGACTTTAGTTTGGGCTCCCAGAACCGTCGAGGGAAAAGAAGTGCAATGGCCAGCGTTGACATTCACACAACTTGCAGCTGACCTGACGATAGAGGCTGACGCCCAGCCAGTGGAATATTTGCATGACAAGCAGCTCCAATTTATGGACTACGCTAATCTACTCCCATATCAGAAATGGGACGCTCACCTCGCCGGTTCAGAGTCCGGTGTCGAGTCCGTTACTCTGTCAGCTCTGTCTGCTGAAGCGTCAGAGTGGCGGACTTCCCAAGACGGCCAGAGAATATTAACCACAACGCCGTCGGTGCTGGGTGGATGTAGAGCTCAGGTATACCGCGCCGCAGGAGCCACTCAGTGGTATACGGCGGTCATCGTTGGAGTCAATGAGCATACGGGGGAGCTGACAGTTACCGATGACACTGTTTTAGAAGAACATAGCGAAGATCCGTCTCTAGTTCAAATGAGGCTTCTCGGTGACGGTGTGATAGAATCGATAATGCGAGGAGAAGTTGTGGGCGTTATGCCGCGGAGATCCCGTTCCAACCTTCAAAGGGTTGAGCGGGAAACAACGAAAAGTCCAGCAACTACTGGAAACAGGAAGAAAACAGGAACCGTTAGGAGCAACGGCCAGATTGCTCAAGTTGAACCTACTCCCCCACCGCCTACAGGATCATCACCGGAACGAGACCCGGCTGAAATTAGCCAAAAAGTGCGATACAATAGTTTTAAAATCATGATGAAACCCACTCAAGGTGCCCTCAAATTCCGGTAA
Protein
MAYKYREDLVGKRFLAVSGLTKINVNRVSEWGWKAGVIRAASLRDNKNKELQVLVEYDGVEWQRREWVAVYSRRTFRVFLVERTLVWAPRTVEGKEVQWPALTFTQLAADLTIEADAQPVEYLHDKQLQFMDYANLLPYQKWDAHLAGSESGVESVTLSALSAEASEWRTSQDGQRILTTTPSVLGGCRAQVYRAAGATQWYTAVIVGVNEHTGELTVTDDTVLEEHSEDPSLVQMRLLGDGVIESIMRGEVVGVMPRRSRSNLQRVERETTKSPATTGNRKKTGTVRSNGQIAQVEPTPPPPTGSSPERDPAEISQKVRYNSFKIMMKPTQGALKFR
Summary
Uniprot
A0A212EH51
A0A2A4JJF8
A0A1B6L5F4
A0A1B6I7W3
A0A1B6JNN6
A0A088AQ66
+ More
A0A139WNI9 A0A139WN32 A0A1B6DJQ3 A0A158ND62 A0A1B6DJN8 A0A0C9R9S2 A0A1Y1LQ77 A0A232FFF7 A0A0A9X2Y6 A0A3L8DQI2 A0A195B5H7 A0A146KUP0 A0A2P2IAS3 A0A0K8T9Y1 A0A0K8T9W6 A0A131ZBQ6 A0A224Z249 T1IIC8 A0A210Q9C8 F4W930 E9I9H7 A0A026WDV4 A0A195F441 A0A151WU44 E2AVL4 A0A195EDX1 L7MJW3 L7MJ95 A0A1S3J155 A0A1S3IZU9 A0A1S3IZB1 A0A131XBM3 A0A154P7C3 A0A0B7BHD0 A0A2C9LIZ7 A0A3P9AA51 A0A3B4CML9 A0A3B4E790 W5MIY4 W5MIW4 A0A1S3QP58 W5UI24 A0A3P8N9M4 A0A2D0S7U7 A0A3P8N9R3 A0A3P9BRP5 A0A3P9BSI4 A0A2D0S7V2 A0A3Q2VGH5 A0A1A8BKQ1 A0A1A8AIU6 A0A3Q0QVL1 A0A3B3R4P4 A0A3B3R5N4 A0A1A8FT17 A0A0Q3REW1 A0A3B4UWK4 A0A2R8QTK3 A0A3B4UWZ5 A0A3Q1FN77 A0A3Q3QJE2 A0A3P8VRI4 A0A3Q3JIP3 A0A3B4ZHM8 A0A3Q1BH92 A0A3Q1CQQ0 A0A0S7GQN2 A0A146VKU5 A0A146ZT90 A0A146VLX7 A0A3Q2NW56 A0A3Q1JG69 A0A3Q1JG76 A0A2K6BIY6 A0A2K5X741 A0A1D5P8D2 F7F8A9 G3UZM1
A0A139WNI9 A0A139WN32 A0A1B6DJQ3 A0A158ND62 A0A1B6DJN8 A0A0C9R9S2 A0A1Y1LQ77 A0A232FFF7 A0A0A9X2Y6 A0A3L8DQI2 A0A195B5H7 A0A146KUP0 A0A2P2IAS3 A0A0K8T9Y1 A0A0K8T9W6 A0A131ZBQ6 A0A224Z249 T1IIC8 A0A210Q9C8 F4W930 E9I9H7 A0A026WDV4 A0A195F441 A0A151WU44 E2AVL4 A0A195EDX1 L7MJW3 L7MJ95 A0A1S3J155 A0A1S3IZU9 A0A1S3IZB1 A0A131XBM3 A0A154P7C3 A0A0B7BHD0 A0A2C9LIZ7 A0A3P9AA51 A0A3B4CML9 A0A3B4E790 W5MIY4 W5MIW4 A0A1S3QP58 W5UI24 A0A3P8N9M4 A0A2D0S7U7 A0A3P8N9R3 A0A3P9BRP5 A0A3P9BSI4 A0A2D0S7V2 A0A3Q2VGH5 A0A1A8BKQ1 A0A1A8AIU6 A0A3Q0QVL1 A0A3B3R4P4 A0A3B3R5N4 A0A1A8FT17 A0A0Q3REW1 A0A3B4UWK4 A0A2R8QTK3 A0A3B4UWZ5 A0A3Q1FN77 A0A3Q3QJE2 A0A3P8VRI4 A0A3Q3JIP3 A0A3B4ZHM8 A0A3Q1BH92 A0A3Q1CQQ0 A0A0S7GQN2 A0A146VKU5 A0A146ZT90 A0A146VLX7 A0A3Q2NW56 A0A3Q1JG69 A0A3Q1JG76 A0A2K6BIY6 A0A2K5X741 A0A1D5P8D2 F7F8A9 G3UZM1
Pubmed
EMBL
AGBW02014956
OWR40817.1
NWSH01001269
PCG71906.1
GEBQ01021028
JAT18949.1
+ More
GECU01024707 JAS82999.1 GECU01006825 JAT00882.1 KQ971311 KYB29437.1 KYB29438.1 GEDC01011423 JAS25875.1 ADTU01000926 ADTU01000927 ADTU01000928 ADTU01000929 ADTU01000930 ADTU01000931 ADTU01000932 ADTU01000933 ADTU01000934 ADTU01000935 GEDC01011453 JAS25845.1 GBYB01003551 JAG73318.1 GEZM01055099 JAV73147.1 NNAY01000343 OXU29047.1 GBHO01029578 GBHO01029574 GDHC01014482 JAG14026.1 JAG14030.1 JAQ04147.1 QOIP01000005 RLU22700.1 KQ976598 KYM79530.1 GDHC01019304 JAP99324.1 IACF01005530 LAB71115.1 GBRD01003477 JAG62344.1 GBRD01003478 JAG62343.1 GEDV01000325 JAP88232.1 GFPF01009497 MAA20643.1 AFFK01014253 NEDP02004518 OWF45352.1 GL888002 EGI69212.1 GL761812 EFZ22768.1 KK107293 EZA53209.1 KQ981855 KYN34937.1 KQ982736 KYQ51430.1 GL443122 EFN62530.1 KQ979039 KYN23306.1 GACK01000759 JAA64275.1 GACK01000758 JAA64276.1 GEFH01003527 JAP65054.1 KQ434829 KZC07777.1 HACG01044861 CEK91726.1 AHAT01004091 AHAT01004092 JT415362 AHH41661.1 HADZ01004261 SBP68202.1 HADY01015680 SBP54165.1 HAEB01014747 SBQ61274.1 LMAW01001203 KQK84129.1 CR354401 CR450773 GBYX01450303 JAO31191.1 GCES01068377 JAR17946.1 GCES01017201 JAR69122.1 GCES01068376 JAR17947.1 AQIA01071610 AQIA01071611 AQIA01071612 AQIA01071613 AQIA01071614 AQIA01071615 AQIA01071616 AQIA01071617 AQIA01071618 AQIA01071619 AADN05000334 JSUE03043769 JSUE03043770 JSUE03043771 JSUE03043772 JSUE03043773 AC155712 AC156272
GECU01024707 JAS82999.1 GECU01006825 JAT00882.1 KQ971311 KYB29437.1 KYB29438.1 GEDC01011423 JAS25875.1 ADTU01000926 ADTU01000927 ADTU01000928 ADTU01000929 ADTU01000930 ADTU01000931 ADTU01000932 ADTU01000933 ADTU01000934 ADTU01000935 GEDC01011453 JAS25845.1 GBYB01003551 JAG73318.1 GEZM01055099 JAV73147.1 NNAY01000343 OXU29047.1 GBHO01029578 GBHO01029574 GDHC01014482 JAG14026.1 JAG14030.1 JAQ04147.1 QOIP01000005 RLU22700.1 KQ976598 KYM79530.1 GDHC01019304 JAP99324.1 IACF01005530 LAB71115.1 GBRD01003477 JAG62344.1 GBRD01003478 JAG62343.1 GEDV01000325 JAP88232.1 GFPF01009497 MAA20643.1 AFFK01014253 NEDP02004518 OWF45352.1 GL888002 EGI69212.1 GL761812 EFZ22768.1 KK107293 EZA53209.1 KQ981855 KYN34937.1 KQ982736 KYQ51430.1 GL443122 EFN62530.1 KQ979039 KYN23306.1 GACK01000759 JAA64275.1 GACK01000758 JAA64276.1 GEFH01003527 JAP65054.1 KQ434829 KZC07777.1 HACG01044861 CEK91726.1 AHAT01004091 AHAT01004092 JT415362 AHH41661.1 HADZ01004261 SBP68202.1 HADY01015680 SBP54165.1 HAEB01014747 SBQ61274.1 LMAW01001203 KQK84129.1 CR354401 CR450773 GBYX01450303 JAO31191.1 GCES01068377 JAR17946.1 GCES01017201 JAR69122.1 GCES01068376 JAR17947.1 AQIA01071610 AQIA01071611 AQIA01071612 AQIA01071613 AQIA01071614 AQIA01071615 AQIA01071616 AQIA01071617 AQIA01071618 AQIA01071619 AADN05000334 JSUE03043769 JSUE03043770 JSUE03043771 JSUE03043772 JSUE03043773 AC155712 AC156272
Proteomes
UP000007151
UP000218220
UP000005203
UP000007266
UP000005205
UP000215335
+ More
UP000279307 UP000078540 UP000242188 UP000007755 UP000053097 UP000078541 UP000075809 UP000000311 UP000078492 UP000085678 UP000076502 UP000076420 UP000265140 UP000261440 UP000018468 UP000087266 UP000221080 UP000265100 UP000265160 UP000264840 UP000261340 UP000261540 UP000051836 UP000261420 UP000000437 UP000257200 UP000261600 UP000265120 UP000261400 UP000257160 UP000265000 UP000265040 UP000233120 UP000233100 UP000000539 UP000006718 UP000000589
UP000279307 UP000078540 UP000242188 UP000007755 UP000053097 UP000078541 UP000075809 UP000000311 UP000078492 UP000085678 UP000076502 UP000076420 UP000265140 UP000261440 UP000018468 UP000087266 UP000221080 UP000265100 UP000265160 UP000264840 UP000261340 UP000261540 UP000051836 UP000261420 UP000000437 UP000257200 UP000261600 UP000265120 UP000261400 UP000257160 UP000265000 UP000265040 UP000233120 UP000233100 UP000000539 UP000006718 UP000000589
PRIDE
Pfam
PF02373 JmjC
Interpro
IPR003347
JmjC_dom
ProteinModelPortal
A0A212EH51
A0A2A4JJF8
A0A1B6L5F4
A0A1B6I7W3
A0A1B6JNN6
A0A088AQ66
+ More
A0A139WNI9 A0A139WN32 A0A1B6DJQ3 A0A158ND62 A0A1B6DJN8 A0A0C9R9S2 A0A1Y1LQ77 A0A232FFF7 A0A0A9X2Y6 A0A3L8DQI2 A0A195B5H7 A0A146KUP0 A0A2P2IAS3 A0A0K8T9Y1 A0A0K8T9W6 A0A131ZBQ6 A0A224Z249 T1IIC8 A0A210Q9C8 F4W930 E9I9H7 A0A026WDV4 A0A195F441 A0A151WU44 E2AVL4 A0A195EDX1 L7MJW3 L7MJ95 A0A1S3J155 A0A1S3IZU9 A0A1S3IZB1 A0A131XBM3 A0A154P7C3 A0A0B7BHD0 A0A2C9LIZ7 A0A3P9AA51 A0A3B4CML9 A0A3B4E790 W5MIY4 W5MIW4 A0A1S3QP58 W5UI24 A0A3P8N9M4 A0A2D0S7U7 A0A3P8N9R3 A0A3P9BRP5 A0A3P9BSI4 A0A2D0S7V2 A0A3Q2VGH5 A0A1A8BKQ1 A0A1A8AIU6 A0A3Q0QVL1 A0A3B3R4P4 A0A3B3R5N4 A0A1A8FT17 A0A0Q3REW1 A0A3B4UWK4 A0A2R8QTK3 A0A3B4UWZ5 A0A3Q1FN77 A0A3Q3QJE2 A0A3P8VRI4 A0A3Q3JIP3 A0A3B4ZHM8 A0A3Q1BH92 A0A3Q1CQQ0 A0A0S7GQN2 A0A146VKU5 A0A146ZT90 A0A146VLX7 A0A3Q2NW56 A0A3Q1JG69 A0A3Q1JG76 A0A2K6BIY6 A0A2K5X741 A0A1D5P8D2 F7F8A9 G3UZM1
A0A139WNI9 A0A139WN32 A0A1B6DJQ3 A0A158ND62 A0A1B6DJN8 A0A0C9R9S2 A0A1Y1LQ77 A0A232FFF7 A0A0A9X2Y6 A0A3L8DQI2 A0A195B5H7 A0A146KUP0 A0A2P2IAS3 A0A0K8T9Y1 A0A0K8T9W6 A0A131ZBQ6 A0A224Z249 T1IIC8 A0A210Q9C8 F4W930 E9I9H7 A0A026WDV4 A0A195F441 A0A151WU44 E2AVL4 A0A195EDX1 L7MJW3 L7MJ95 A0A1S3J155 A0A1S3IZU9 A0A1S3IZB1 A0A131XBM3 A0A154P7C3 A0A0B7BHD0 A0A2C9LIZ7 A0A3P9AA51 A0A3B4CML9 A0A3B4E790 W5MIY4 W5MIW4 A0A1S3QP58 W5UI24 A0A3P8N9M4 A0A2D0S7U7 A0A3P8N9R3 A0A3P9BRP5 A0A3P9BSI4 A0A2D0S7V2 A0A3Q2VGH5 A0A1A8BKQ1 A0A1A8AIU6 A0A3Q0QVL1 A0A3B3R4P4 A0A3B3R5N4 A0A1A8FT17 A0A0Q3REW1 A0A3B4UWK4 A0A2R8QTK3 A0A3B4UWZ5 A0A3Q1FN77 A0A3Q3QJE2 A0A3P8VRI4 A0A3Q3JIP3 A0A3B4ZHM8 A0A3Q1BH92 A0A3Q1CQQ0 A0A0S7GQN2 A0A146VKU5 A0A146ZT90 A0A146VLX7 A0A3Q2NW56 A0A3Q1JG69 A0A3Q1JG76 A0A2K6BIY6 A0A2K5X741 A0A1D5P8D2 F7F8A9 G3UZM1
Ontologies
GO
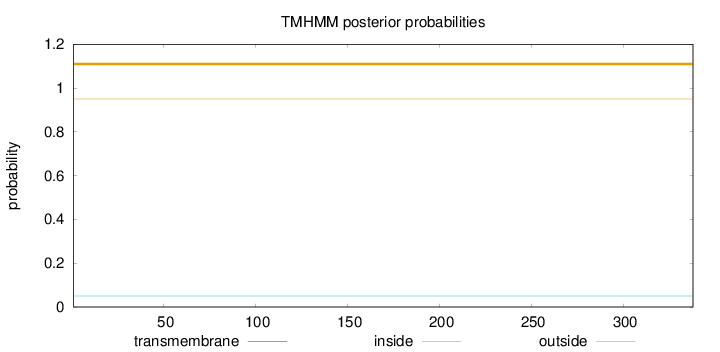
Topology
Length:
338
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00385000000000001
Exp number, first 60 AAs:
0.00228
Total prob of N-in:
0.05007
outside
1 - 338
Population Genetic Test Statistics
Pi
238.93656
Theta
194.633999
Tajima's D
0.763295
CLR
0.282183
CSRT
0.591420428978551
Interpretation
Uncertain
