Pre Gene Modal
BGIBMGA011568
Annotation
Testis-expressed_sequence_10_protein-like_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.641
Sequence
CDS
ATGCCACAAACGGGTGCAACTCGTTACCAAAAATTTCTTAAGTCTGAAAAGGCAAAGACTAAACTGAAAGCCAAAAAGGATCTTCCTAAAGGAACAAATGTTACTAAAACAAATTTTAAAGTGAAGAAAATTGTCATTAAAGAGCAGCTTAGAAAACGTACTGAGAAAGAGGCGTTGTCTATACGAAAGCTGAATGTTAAAGAACTCATAACACGTCTAAATCATTTTAACACAAATACCCGGAAAGACGCTCTCGATGGACTCACGGAACTAATTTCAGCTCATCCGGAAATTTTAGAAAAAGATTTAGGACTAATTATTCATGGAGTTAGTCCAATGATTTTGAATGTTGAAAAAATCGTGCGCCATGAATCACTGAAAGTTTTACACTTGATACTTTCGAATTTGAGTGTTGAGAAGATAGATCCATTTTTTGATGTTATGTCTACGTTTCTAAGGAGTGCTATGACTCATATTAACAGTAGAATTCAAGAAGATTCTCTACTATTCTTAGATATTCTCCTTCTATGTGCCCCAATGAAAATGGCAAAAGATTTTTATAGAATTATTCCAAATTTTTTAGACATGATATCAAAATTACACATTGACTCAAAACCTGGACGAACACTTACAGTTAATATGGAAAGCCAAATTACAAGTGTTCGTTGGAGATCTAAAGTTCTTCATAGACTTCAAGATTATTTACAGAAATTTGTCATACACAACAAAGTTCTTGAATTTGAGAAACCTATAACAGAGTGTACAGAAGTTTTTGACCAAACTAAGCTAAAGTATTATCCATTATTTAAGCCAACTTACACATCAGTATGCTATTTGAAAATGCTATCATCCAAAAATTCTCAAGAGATTGTATCTGTTGATGAAGTGGAACAATTGAAAGGCTATATAGACAACTTGATGCCACTATTATTTGAAATATGGCTTGAAGTGTGCCCAAATATGAACAAGGAAAAAAACATTGAGACTGTTGTAAGTGAAGATGCTGCATCATTATTAAAACATTCTCTTGAGATTATATTTTTGTTATGGGACCTAATAGAACATCTCAACAAGAAGAATCCGAGTTCAGATATAGAATCATCTTTTTGCCAGAAATATAAAAACTTATATACATTACACTTTGTGAATGCTTTTCCTTATGTGACAAATGTTAGAACCAAACAAAATAGTGATACTTCCCTTGAGAATGTGATTACGGACTCAAAATTGGTTTTAGAAAATTTGAAAATATGCCACTTGTTCATATTATTAAATCCAAAGATAAATTTGAAGTTACAAAACAAAGACATTGTTGCTATCTTGCATTATATAGAAAAAACTTTCAACCATAATACTAAAGATGAAGTAAACGATGCCATCATTGAAATCATGCACACCATATTTTCAACTGAAGTGAGTAACTGGAGTAAAACATGTAGTGTAATGGATGGCTTATTCCGGAAAATTATATGTGCCTACTTCAATAAAAATATGTCCGGATCGTGGAAACAAAAAATATTCGGGGTCCTATGCAAAGTTGCTCTTAATGATAAACTAAGTCATTTTCATGCCATTGATGCATATGAAATGTGGCTCAAGAATCTACCAGACATTTTATTGGGTAGTTCAGTTACAGAACAAACCATAGATATTATTCATAAATTTGCTGTACAGAGCAATGACAAATTTAATATAGTCATCAAACCTAAGTTGTCAATGATTATAGAAAATCTGCCAAATATTGTTAGTTCAGAAGCTGCTAATAATCCAAACATGTATTATAAATTATTTTCATTATTTTTTTGGATACGACCTTGGGATAATGAATCCTTAAATGTTTTAGAAAAACAATTATTGGACAGCAAATATAGTGATGATCATTGTAAATATATTATTGATACGTTAAAAAGTAGAACTGGAAGCACCTAA
Protein
MPQTGATRYQKFLKSEKAKTKLKAKKDLPKGTNVTKTNFKVKKIVIKEQLRKRTEKEALSIRKLNVKELITRLNHFNTNTRKDALDGLTELISAHPEILEKDLGLIIHGVSPMILNVEKIVRHESLKVLHLILSNLSVEKIDPFFDVMSTFLRSAMTHINSRIQEDSLLFLDILLLCAPMKMAKDFYRIIPNFLDMISKLHIDSKPGRTLTVNMESQITSVRWRSKVLHRLQDYLQKFVIHNKVLEFEKPITECTEVFDQTKLKYYPLFKPTYTSVCYLKMLSSKNSQEIVSVDEVEQLKGYIDNLMPLLFEIWLEVCPNMNKEKNIETVVSEDAASLLKHSLEIIFLLWDLIEHLNKKNPSSDIESSFCQKYKNLYTLHFVNAFPYVTNVRTKQNSDTSLENVITDSKLVLENLKICHLFILLNPKINLKLQNKDIVAILHYIEKTFNHNTKDEVNDAIIEIMHTIFSTEVSNWSKTCSVMDGLFRKIICAYFNKNMSGSWKQKIFGVLCKVALNDKLSHFHAIDAYEMWLKNLPDILLGSSVTEQTIDIIHKFAVQSNDKFNIVIKPKLSMIIENLPNIVSSEAANNPNMYYKLFSLFFWIRPWDNESLNVLEKQLLDSKYSDDHCKYIIDTLKSRTGST
Summary
Uniprot
H9JPW3
A0A2H1V7T9
A0A0L7LHL2
A0A2A4IXU1
A0A1E1WRU8
A0A194QTA1
+ More
A0A212EH72 A0A194PSQ7 K7IWV6 A0A2J7RRZ2 A0A232FCM9 A0A182JFZ7 A0A1Y1NA26 A0A088AR13 Q17MP9 A0A182G568 A0A1Q3EWC5 A0A1I8NIA4 A0A2J7RRZ4 A0A0L0C4I3 Q5TSF1 A0A182S5U2 U5ENT9 A0A182S4X7 A0A2A3EAF6 A0A084VKW5 A0A182LQ17 A0A182YA36 A0A182HFR1 A0A182MC05 A0A182JRK6 A0A182UR51 A0A182VTC1 A0A0V0G6P4 A0A1A9UUW6 E2A6T9 A0A182X1U7 A0A0A1WDU1 A0A023EXT6 A0A182P8H0 A0A1B0FCU1 A0A1A9XYF2 A0A034VSP7 A0A182R137 A0A195FLP2 A0A1A9XYE9 A0A1B0ASW0 A0A336MCB0 F4W5R0 A0A1I8PX04 A0A0K8VNE7 A0A1A9WZE0 A0A0C9RY26 A0A151I0Q5 A0A158P2Q7 W8AX17 A0A1B6D5G6 N6TVW5 T1I686 A0A146M6D5 A0A146LN70 A0A1B6KGZ3 A0A1B6L904 D2A610 B4Q0M8 A0A1W4VWK5 B4R6A8 B4L8I3 A0A1B6JIM9 A0A0C9QEE7 A0A0M5J5Y3 B4IKW6 C3Z8G3 Q9W3K4 Q29HE4 B4GXV1 E9J5D5 B3MRK3 B3NWJ9 A0A3B0K3U1 B4M1Y6 B4NPM9 A0A2U9AYK4 A0A2H8TGP5 A0A3Q3E887 J9JZN0 A0A091NU67 A0A3Q2ZTR3
A0A212EH72 A0A194PSQ7 K7IWV6 A0A2J7RRZ2 A0A232FCM9 A0A182JFZ7 A0A1Y1NA26 A0A088AR13 Q17MP9 A0A182G568 A0A1Q3EWC5 A0A1I8NIA4 A0A2J7RRZ4 A0A0L0C4I3 Q5TSF1 A0A182S5U2 U5ENT9 A0A182S4X7 A0A2A3EAF6 A0A084VKW5 A0A182LQ17 A0A182YA36 A0A182HFR1 A0A182MC05 A0A182JRK6 A0A182UR51 A0A182VTC1 A0A0V0G6P4 A0A1A9UUW6 E2A6T9 A0A182X1U7 A0A0A1WDU1 A0A023EXT6 A0A182P8H0 A0A1B0FCU1 A0A1A9XYF2 A0A034VSP7 A0A182R137 A0A195FLP2 A0A1A9XYE9 A0A1B0ASW0 A0A336MCB0 F4W5R0 A0A1I8PX04 A0A0K8VNE7 A0A1A9WZE0 A0A0C9RY26 A0A151I0Q5 A0A158P2Q7 W8AX17 A0A1B6D5G6 N6TVW5 T1I686 A0A146M6D5 A0A146LN70 A0A1B6KGZ3 A0A1B6L904 D2A610 B4Q0M8 A0A1W4VWK5 B4R6A8 B4L8I3 A0A1B6JIM9 A0A0C9QEE7 A0A0M5J5Y3 B4IKW6 C3Z8G3 Q9W3K4 Q29HE4 B4GXV1 E9J5D5 B3MRK3 B3NWJ9 A0A3B0K3U1 B4M1Y6 B4NPM9 A0A2U9AYK4 A0A2H8TGP5 A0A3Q3E887 J9JZN0 A0A091NU67 A0A3Q2ZTR3
Pubmed
19121390
26227816
26354079
22118469
20075255
28648823
+ More
28004739 17510324 26483478 25315136 26108605 12364791 24438588 20966253 25244985 20798317 25830018 25474469 25348373 21719571 21347285 24495485 23537049 26823975 18362917 19820115 17994087 17550304 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665
28004739 17510324 26483478 25315136 26108605 12364791 24438588 20966253 25244985 20798317 25830018 25474469 25348373 21719571 21347285 24495485 23537049 26823975 18362917 19820115 17994087 17550304 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665
EMBL
BABH01015440
ODYU01001116
SOQ36881.1
JTDY01001078
KOB74942.1
NWSH01004849
+ More
PCG64655.1 GDQN01001352 JAT89702.1 KQ461195 KPJ06771.1 AGBW02014956 OWR40820.1 KQ459600 KPI94160.1 AAZX01010983 NEVH01000596 PNF43611.1 NNAY01000459 OXU28243.1 GEZM01009708 JAV94368.1 CH477204 EAT47973.1 JXUM01145173 JXUM01145174 JXUM01145175 JXUM01145176 JXUM01145177 KQ569935 KXJ68480.1 GFDL01015440 JAV19605.1 PNF43613.1 JRES01000994 KNC26354.1 AAAB01008933 EAL40354.3 GANO01003941 JAB55930.1 KZ288308 PBC28743.1 ATLV01014310 KE524956 KFB38609.1 APCN01005566 AXCM01002293 GECL01002517 JAP03607.1 GL437203 EFN70805.1 GBXI01017471 JAC96820.1 GBBI01004622 JAC14090.1 CCAG010007386 GAKP01013463 JAC45489.1 AXCN02001048 KQ981512 KYN40909.1 JXJN01003016 UFQS01000641 UFQT01000641 SSX05668.1 SSX26027.1 GL887695 EGI70386.1 GDHF01011908 JAI40406.1 GBYB01012800 GBYB01012801 JAG82567.1 JAG82568.1 KQ976609 KYM79316.1 ADTU01007382 GAMC01013090 JAB93465.1 GEDC01031005 GEDC01016367 JAS06293.1 JAS20931.1 APGK01052898 KB741216 ENN72526.1 ACPB03014449 GDHC01004313 JAQ14316.1 GDHC01010397 JAQ08232.1 GEBQ01029271 JAT10706.1 GEBQ01019784 JAT20193.1 KQ971346 EFA04998.2 CM000162 EDX02299.2 CM000366 EDX17385.1 CH933815 EDW07958.1 GECU01020525 GECU01017978 GECU01008658 JAS87181.1 JAS89728.1 JAS99048.1 GBYB01012803 JAG82570.1 CP012528 ALC48656.1 CH480859 EDW52737.1 GG666594 EEN51068.1 AE014298 AY061490 AAF46322.1 AAL29038.1 AHN59474.1 CH379064 EAL31814.1 CH479196 EDW27578.1 GL768143 EFZ11969.1 CH902622 EDV34408.1 CH954180 EDV46819.1 OUUW01000016 SPP88897.1 CH940651 EDW65690.2 CH964291 EDW86469.1 CP026243 AWO96641.1 GFXV01001405 MBW13210.1 ABLF02035128 ABLF02035132 KL393535 KFP92484.1
PCG64655.1 GDQN01001352 JAT89702.1 KQ461195 KPJ06771.1 AGBW02014956 OWR40820.1 KQ459600 KPI94160.1 AAZX01010983 NEVH01000596 PNF43611.1 NNAY01000459 OXU28243.1 GEZM01009708 JAV94368.1 CH477204 EAT47973.1 JXUM01145173 JXUM01145174 JXUM01145175 JXUM01145176 JXUM01145177 KQ569935 KXJ68480.1 GFDL01015440 JAV19605.1 PNF43613.1 JRES01000994 KNC26354.1 AAAB01008933 EAL40354.3 GANO01003941 JAB55930.1 KZ288308 PBC28743.1 ATLV01014310 KE524956 KFB38609.1 APCN01005566 AXCM01002293 GECL01002517 JAP03607.1 GL437203 EFN70805.1 GBXI01017471 JAC96820.1 GBBI01004622 JAC14090.1 CCAG010007386 GAKP01013463 JAC45489.1 AXCN02001048 KQ981512 KYN40909.1 JXJN01003016 UFQS01000641 UFQT01000641 SSX05668.1 SSX26027.1 GL887695 EGI70386.1 GDHF01011908 JAI40406.1 GBYB01012800 GBYB01012801 JAG82567.1 JAG82568.1 KQ976609 KYM79316.1 ADTU01007382 GAMC01013090 JAB93465.1 GEDC01031005 GEDC01016367 JAS06293.1 JAS20931.1 APGK01052898 KB741216 ENN72526.1 ACPB03014449 GDHC01004313 JAQ14316.1 GDHC01010397 JAQ08232.1 GEBQ01029271 JAT10706.1 GEBQ01019784 JAT20193.1 KQ971346 EFA04998.2 CM000162 EDX02299.2 CM000366 EDX17385.1 CH933815 EDW07958.1 GECU01020525 GECU01017978 GECU01008658 JAS87181.1 JAS89728.1 JAS99048.1 GBYB01012803 JAG82570.1 CP012528 ALC48656.1 CH480859 EDW52737.1 GG666594 EEN51068.1 AE014298 AY061490 AAF46322.1 AAL29038.1 AHN59474.1 CH379064 EAL31814.1 CH479196 EDW27578.1 GL768143 EFZ11969.1 CH902622 EDV34408.1 CH954180 EDV46819.1 OUUW01000016 SPP88897.1 CH940651 EDW65690.2 CH964291 EDW86469.1 CP026243 AWO96641.1 GFXV01001405 MBW13210.1 ABLF02035128 ABLF02035132 KL393535 KFP92484.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000002358 UP000235965 UP000215335 UP000075880 UP000005203 UP000008820 UP000069940 UP000249989 UP000095301 UP000037069 UP000007062 UP000075901 UP000075900 UP000242457 UP000030765 UP000075882 UP000076408 UP000075840 UP000075883 UP000075881 UP000075903 UP000075920 UP000078200 UP000000311 UP000076407 UP000075885 UP000092444 UP000092443 UP000075886 UP000078541 UP000092460 UP000007755 UP000095300 UP000091820 UP000078540 UP000005205 UP000019118 UP000015103 UP000007266 UP000002282 UP000192221 UP000000304 UP000009192 UP000092553 UP000001292 UP000001554 UP000000803 UP000001819 UP000008744 UP000007801 UP000008711 UP000268350 UP000008792 UP000007798 UP000246464 UP000261660 UP000007819 UP000264800
UP000002358 UP000235965 UP000215335 UP000075880 UP000005203 UP000008820 UP000069940 UP000249989 UP000095301 UP000037069 UP000007062 UP000075901 UP000075900 UP000242457 UP000030765 UP000075882 UP000076408 UP000075840 UP000075883 UP000075881 UP000075903 UP000075920 UP000078200 UP000000311 UP000076407 UP000075885 UP000092444 UP000092443 UP000075886 UP000078541 UP000092460 UP000007755 UP000095300 UP000091820 UP000078540 UP000005205 UP000019118 UP000015103 UP000007266 UP000002282 UP000192221 UP000000304 UP000009192 UP000092553 UP000001292 UP000001554 UP000000803 UP000001819 UP000008744 UP000007801 UP000008711 UP000268350 UP000008792 UP000007798 UP000246464 UP000261660 UP000007819 UP000264800
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JPW3
A0A2H1V7T9
A0A0L7LHL2
A0A2A4IXU1
A0A1E1WRU8
A0A194QTA1
+ More
A0A212EH72 A0A194PSQ7 K7IWV6 A0A2J7RRZ2 A0A232FCM9 A0A182JFZ7 A0A1Y1NA26 A0A088AR13 Q17MP9 A0A182G568 A0A1Q3EWC5 A0A1I8NIA4 A0A2J7RRZ4 A0A0L0C4I3 Q5TSF1 A0A182S5U2 U5ENT9 A0A182S4X7 A0A2A3EAF6 A0A084VKW5 A0A182LQ17 A0A182YA36 A0A182HFR1 A0A182MC05 A0A182JRK6 A0A182UR51 A0A182VTC1 A0A0V0G6P4 A0A1A9UUW6 E2A6T9 A0A182X1U7 A0A0A1WDU1 A0A023EXT6 A0A182P8H0 A0A1B0FCU1 A0A1A9XYF2 A0A034VSP7 A0A182R137 A0A195FLP2 A0A1A9XYE9 A0A1B0ASW0 A0A336MCB0 F4W5R0 A0A1I8PX04 A0A0K8VNE7 A0A1A9WZE0 A0A0C9RY26 A0A151I0Q5 A0A158P2Q7 W8AX17 A0A1B6D5G6 N6TVW5 T1I686 A0A146M6D5 A0A146LN70 A0A1B6KGZ3 A0A1B6L904 D2A610 B4Q0M8 A0A1W4VWK5 B4R6A8 B4L8I3 A0A1B6JIM9 A0A0C9QEE7 A0A0M5J5Y3 B4IKW6 C3Z8G3 Q9W3K4 Q29HE4 B4GXV1 E9J5D5 B3MRK3 B3NWJ9 A0A3B0K3U1 B4M1Y6 B4NPM9 A0A2U9AYK4 A0A2H8TGP5 A0A3Q3E887 J9JZN0 A0A091NU67 A0A3Q2ZTR3
A0A212EH72 A0A194PSQ7 K7IWV6 A0A2J7RRZ2 A0A232FCM9 A0A182JFZ7 A0A1Y1NA26 A0A088AR13 Q17MP9 A0A182G568 A0A1Q3EWC5 A0A1I8NIA4 A0A2J7RRZ4 A0A0L0C4I3 Q5TSF1 A0A182S5U2 U5ENT9 A0A182S4X7 A0A2A3EAF6 A0A084VKW5 A0A182LQ17 A0A182YA36 A0A182HFR1 A0A182MC05 A0A182JRK6 A0A182UR51 A0A182VTC1 A0A0V0G6P4 A0A1A9UUW6 E2A6T9 A0A182X1U7 A0A0A1WDU1 A0A023EXT6 A0A182P8H0 A0A1B0FCU1 A0A1A9XYF2 A0A034VSP7 A0A182R137 A0A195FLP2 A0A1A9XYE9 A0A1B0ASW0 A0A336MCB0 F4W5R0 A0A1I8PX04 A0A0K8VNE7 A0A1A9WZE0 A0A0C9RY26 A0A151I0Q5 A0A158P2Q7 W8AX17 A0A1B6D5G6 N6TVW5 T1I686 A0A146M6D5 A0A146LN70 A0A1B6KGZ3 A0A1B6L904 D2A610 B4Q0M8 A0A1W4VWK5 B4R6A8 B4L8I3 A0A1B6JIM9 A0A0C9QEE7 A0A0M5J5Y3 B4IKW6 C3Z8G3 Q9W3K4 Q29HE4 B4GXV1 E9J5D5 B3MRK3 B3NWJ9 A0A3B0K3U1 B4M1Y6 B4NPM9 A0A2U9AYK4 A0A2H8TGP5 A0A3Q3E887 J9JZN0 A0A091NU67 A0A3Q2ZTR3
Ontologies
KEGG
GO
PANTHER
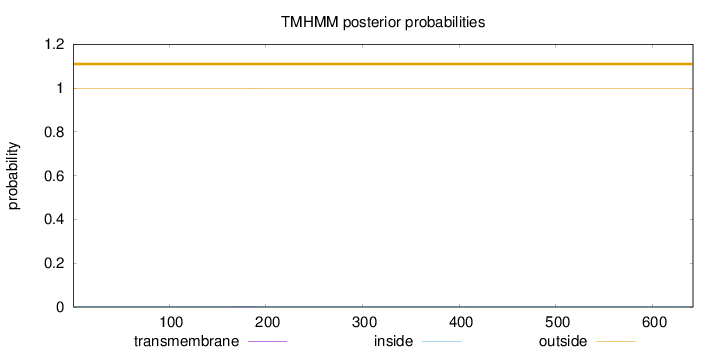
Topology
Length:
642
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0460700000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00348
outside
1 - 642
Population Genetic Test Statistics
Pi
233.711955
Theta
193.894415
Tajima's D
0.764433
CLR
0.073919
CSRT
0.594120293985301
Interpretation
Uncertain
