Gene
KWMTBOMO13840
Annotation
protein_tyrosine_phosphatase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.68
Sequence
CDS
ATGGAAATCCGCATTGTTAGTCAACAAGTTCAAAAATCACAGGTGAAAAAAATATGGGACATTCCGGCGCTTCTAGAGGCGATACCCAAACTCGGTGCAGTCATAGATTTGACAAATACCGATAAATATTATAAACCTGAGGATGTAAAGGCTGCAGGTATTTTACATAAAAAAATCATAATGCCCGGTCGAATTTTGCCACCCGAAAACAAAGTGAAAGAATTTATGGATGCCGTCGATGATTTCCTGGGAAAAGACTCCGATATCCTGTTAGGGGTGCACTGCACGCACGGATTAAACCGCACGGGTTATATGGTGTGCCGATATATGAGGGATCGCCTCGGAATGTCGGGAAAGGAGGCTATTAAAAAATTCGAAAGAGCACGTGGCTATGCCATTGAAAGGATAGTTTACACATCCGATTTATTGAAGCAAAATCCCGCTATTGACAAAGAGCGGACTAACAGCAAATCGGCTACAAAGAGACGTCACGACTTTGAATACAGATCACCTTTAGCTGAAGAGTACGAAACTGAAACAACTAGAAGAAGGTATCGTTAG
Protein
MEIRIVSQQVQKSQVKKIWDIPALLEAIPKLGAVIDLTNTDKYYKPEDVKAAGILHKKIIMPGRILPPENKVKEFMDAVDDFLGKDSDILLGVHCTHGLNRTGYMVCRYMRDRLGMSGKEAIKKFERARGYAIERIVYTSDLLKQNPAIDKERTNSKSATKRRHDFEYRSPLAEEYETETTRRRYR
Summary
Uniprot
Q75T82
A0A139WNF3
Q2NP56
A0A2I7MM49
A1YRI5
A0A1A9VMC7
+ More
A0A1B0BAL7 A0A0P8Y5F1 A0A0P8ZQI0 A0A1L8DGA9 A0A0K2V4B8 A0A1A9ZP31 B4LIV8 A0A1B0F993 B3MCW3 A0A1W4UZP0 B4KUG1 A0A0K2V5G6 A0A1I8N9A8 A0A1I8NXI8 B4P619 Q8SX38 A0A1S3JS80 J3JYW8 A0A1Y1M8E9 B4HNS5 A0A1E1X2L3 A0A3B0J5L2 A0A0J9TZ56 A0A224YYK9 A0A0J9RBH7 A0A3L8E008 A0A0P4YLH4
A0A1B0BAL7 A0A0P8Y5F1 A0A0P8ZQI0 A0A1L8DGA9 A0A0K2V4B8 A0A1A9ZP31 B4LIV8 A0A1B0F993 B3MCW3 A0A1W4UZP0 B4KUG1 A0A0K2V5G6 A0A1I8N9A8 A0A1I8NXI8 B4P619 Q8SX38 A0A1S3JS80 J3JYW8 A0A1Y1M8E9 B4HNS5 A0A1E1X2L3 A0A3B0J5L2 A0A0J9TZ56 A0A224YYK9 A0A0J9RBH7 A0A3L8E008 A0A0P4YLH4
Pubmed
EMBL
AB126695
BAD02367.1
KQ971312
KYB29396.1
AP009046
BAE72431.1
+ More
KY550224 AUR45159.1 EF125867 ABM05439.1 JXJN01011052 CH902619 KPU76754.1 KPU76753.1 GFDF01008586 JAV05498.1 HACA01028007 CDW45368.1 CH940648 EDW60408.2 CCAG010021392 EDV37365.1 CH933808 EDW10748.2 HACA01028006 CDW45367.1 CM000158 EDW90894.1 AE013599 AY094864 AAF58626.2 AAM11217.1 BT128449 AEE63406.1 GEZM01037460 JAV82059.1 CH480816 EDW47440.1 GFAC01005691 JAT93497.1 OUUW01000001 SPP74962.1 CM002911 KMY93030.1 GFPF01011540 MAA22686.1 KMY93029.1 QOIP01000002 RLU25946.1 GDIP01225882 JAI97519.1
KY550224 AUR45159.1 EF125867 ABM05439.1 JXJN01011052 CH902619 KPU76754.1 KPU76753.1 GFDF01008586 JAV05498.1 HACA01028007 CDW45368.1 CH940648 EDW60408.2 CCAG010021392 EDV37365.1 CH933808 EDW10748.2 HACA01028006 CDW45367.1 CM000158 EDW90894.1 AE013599 AY094864 AAF58626.2 AAM11217.1 BT128449 AEE63406.1 GEZM01037460 JAV82059.1 CH480816 EDW47440.1 GFAC01005691 JAT93497.1 OUUW01000001 SPP74962.1 CM002911 KMY93030.1 GFPF01011540 MAA22686.1 KMY93029.1 QOIP01000002 RLU25946.1 GDIP01225882 JAI97519.1
Proteomes
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
Q75T82
A0A139WNF3
Q2NP56
A0A2I7MM49
A1YRI5
A0A1A9VMC7
+ More
A0A1B0BAL7 A0A0P8Y5F1 A0A0P8ZQI0 A0A1L8DGA9 A0A0K2V4B8 A0A1A9ZP31 B4LIV8 A0A1B0F993 B3MCW3 A0A1W4UZP0 B4KUG1 A0A0K2V5G6 A0A1I8N9A8 A0A1I8NXI8 B4P619 Q8SX38 A0A1S3JS80 J3JYW8 A0A1Y1M8E9 B4HNS5 A0A1E1X2L3 A0A3B0J5L2 A0A0J9TZ56 A0A224YYK9 A0A0J9RBH7 A0A3L8E008 A0A0P4YLH4
A0A1B0BAL7 A0A0P8Y5F1 A0A0P8ZQI0 A0A1L8DGA9 A0A0K2V4B8 A0A1A9ZP31 B4LIV8 A0A1B0F993 B3MCW3 A0A1W4UZP0 B4KUG1 A0A0K2V5G6 A0A1I8N9A8 A0A1I8NXI8 B4P619 Q8SX38 A0A1S3JS80 J3JYW8 A0A1Y1M8E9 B4HNS5 A0A1E1X2L3 A0A3B0J5L2 A0A0J9TZ56 A0A224YYK9 A0A0J9RBH7 A0A3L8E008 A0A0P4YLH4
PDB
1YN9
E-value=7.11376e-36,
Score=373
Ontologies
GO
Topology
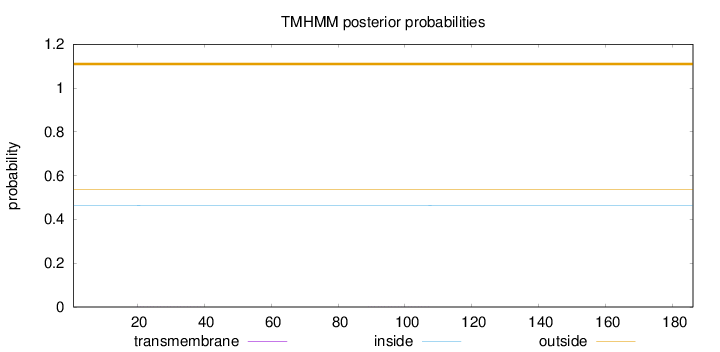
Length:
186
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01146
Exp number, first 60 AAs:
0.00676
Total prob of N-in:
0.46441
outside
1 - 186
Population Genetic Test Statistics
Pi
233.992756
Theta
183.976843
Tajima's D
0.990126
CLR
0.344758
CSRT
0.655817209139543
Interpretation
Uncertain
