Gene
KWMTBOMO13836
Annotation
Suppressor_of_tumorigenicity_14_protein-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.846 PlasmaMembrane Reliability : 1.322
Sequence
CDS
ATGTTTAAATTACTAAGTAAATTAAGCATTTTATTATCATTTATACAGTTGTGTGTTTCTATGAATAGTTTTTTGTATTTTAATAAGAGCGAGTTCGTGCAGAGGGTGTCTAATGGTAGAGCAGCCAAACTCGGAGATGTACCGTATCAGGTTGCTTTTAAAGCTTTGTATTCAAGAATTCGTAATTTGTACGTCACCTTCTGCGGTGGTGTGATAATAGGGCCTGCGAAGCTGATCTCTGCTGCACATTGCTTTGAAGAAAAAAATTCTTACTGTCGGAAGTTTTGGAGTAAGAGGTAA
Protein
MFKLLSKLSILLSFIQLCVSMNSFLYFNKSEFVQRVSNGRAAKLGDVPYQVAFKALYSRIRNLYVTFCGGVIIGPAKLISAAHCFEEKNSYCRKFWSKR
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
Interpro
CDD
ProteinModelPortal
PDB
2XRC
E-value=2.61871e-05,
Score=107
Ontologies
GO
Topology
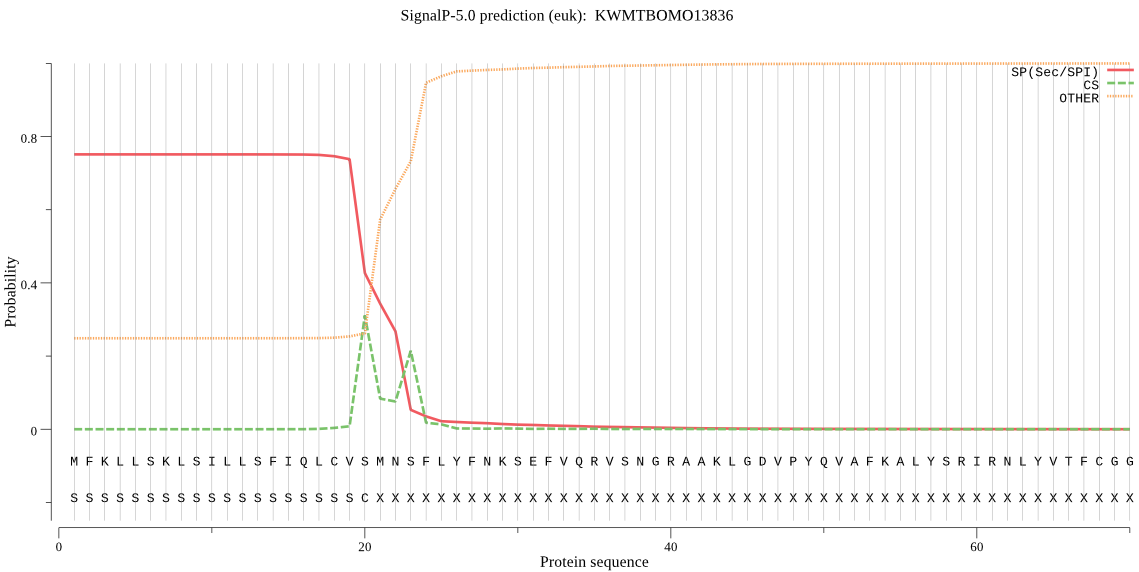
SignalP
Position: 1 - 20,
Likelihood: 0.750829
Length:
99
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
36.72583
Exp number, first 60 AAs:
17.16971
Total prob of N-in:
0.29721
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 27
inside
28 - 62
TMhelix
63 - 85
outside
86 - 99

Population Genetic Test Statistics
Pi
237.616739
Theta
168.039503
Tajima's D
1.20225
CLR
27.478718
CSRT
0.71656417179141
Interpretation
Uncertain
