Gene
KWMTBOMO13835
Pre Gene Modal
BGIBMGA011563
Annotation
(11Z)-hexadec-11-enoyl-CoA_conjugase_[Bombyx_mori]
Full name
(11Z)-hexadec-11-enoyl-CoA conjugase
+ More
Acyl-CoA Delta(11) desaturase
Acyl-CoA Delta(11) desaturase
Alternative Name
Acyl-CoA Delta(11) desaturase
Acyl-CoA Z11/delta10,12 desaturase
Acyl-CoA Delta-11 desaturase
Acyl-CoA Z/E11 desaturase
SlsZ/E11
Acyl-CoA Z11/delta10,12 desaturase
Acyl-CoA Delta-11 desaturase
Acyl-CoA Z/E11 desaturase
SlsZ/E11
Location in the cell
PlasmaMembrane Reliability : 3.466
Sequence
CDS
ATGCCTCCTAATTCAGTGGATAAAACAAATGAAACAGAATACCTCAAGGATAATCATGTTGATTATGAAAAACTCATTGCACCCCAGGCCTCGCCCATCAAACACAAAATTGTAGTGATGAACGTGATACGTTTTAGCTATTTACATATTGCTGGTCTATATGGACTTTATCTCTGTTTCACCTCAGCAAAATTGGCTACATCGGTTTTTGCTATTGTGTTATTCTTCCTTGGGAACTTTGGTATTACAGCTGGAGCTCATCGTCTGTGGTCTCATAATGGTTACAAAGTCAAACTGCCCCTCGAGATCCTGTTGATGGTCTTCAACAGTATTGCTTTTCAGAACACCATTTTCACATGGGTGAGAGATCACAGGCTACATCACAAGTATACGGACACTGATGCAGACCCTCATAATGCTACGAGAGGTTTTTTTTTCTCACACATCGGCTGGTTGTTGGTGCGAAAACATCCAATGGTGAAAATTGCTGGAAAGAGCCTTGACATGTCAGATATTTATTGTAATCCTCTTTTAAGATTTCAGAAAAAGTACGCAATACCATTTATAGGAACAATATGTTTTATCATACCGACATTAGCTCCAATGTATTTTTGGGGTGAAAGCTTAAACAACGCTTGGCATATTACAGTTTTAAGATACATTTTCAGTCTCAATGGCACTTTTCTTGTAAATAGCGCTGCTCATTTGTGGGGATACAAACCGTACGATAAGAGCCTGAAAGCTACTCAAAGTGGCATGGCAAATGCATTTACTTTTGGTGAAGGATTTCACAATTACCATCACGTTTTTCCGTGGGATTATCGGGCAGACGAATTGGGGGACAGGTACATAAATTTGACAACAAGGTTCATAGATTTCTTTGCATGGATGGGATGGGCTTATGATCTGAAAACTGCTTCCACCAATATTATTGAGAAGAGGGCACTAAGAACAGGGGATGGGACTTACAAACGACCAAATGGAATGAATTGA
Protein
MPPNSVDKTNETEYLKDNHVDYEKLIAPQASPIKHKIVVMNVIRFSYLHIAGLYGLYLCFTSAKLATSVFAIVLFFLGNFGITAGAHRLWSHNGYKVKLPLEILLMVFNSIAFQNTIFTWVRDHRLHHKYTDTDADPHNATRGFFFSHIGWLLVRKHPMVKIAGKSLDMSDIYCNPLLRFQKKYAIPFIGTICFIIPTLAPMYFWGESLNNAWHITVLRYIFSLNGTFLVNSAAHLWGYKPYDKSLKATQSGMANAFTFGEGFHNYHHVFPWDYRADELGDRYINLTTRFIDFFAWMGWAYDLKTASTNIIEKRALRTGDGTYKRPNGMN
Summary
Description
Fatty acid desaturase that catalyzes 2 consecutive steps in the biosynthesis of bombykol, a sex pheromone produced by the moth. First acts as an acyl-CoA Delta(11) desaturase (1) by catalyzing the formation of Delta(11) fatty acyl precursors. Then acts as a (11Z)-hexadec-11-enoyl-CoA conjugase (2) by converting a single cis double bond at position 11 of (11Z)-hexadec-11-enoyl-CoA into conjugated 10 trans and 12 cis double bonds.
Catalyzes the formation of Delta(11) fatty acyl precursors in the pheromone gland, with a preference for myristic acid.
Catalyzes the formation of delta(11) fatty acyl precursors in the pheromone gland, and has high activity towards palmitic acid and stearic acid.
Catalyzes the formation of Delta(11) fatty acyl precursors in the pheromone gland.
Catalyzes the formation of Delta(11) fatty acyl precursors in the pheromone gland, with a preference for myristic acid.
Catalyzes the formation of delta(11) fatty acyl precursors in the pheromone gland, and has high activity towards palmitic acid and stearic acid.
Catalyzes the formation of Delta(11) fatty acyl precursors in the pheromone gland.
Catalytic Activity
2 [Fe(II)-cytochrome b5] + an 11,12-saturated fatty acyl-CoA + 2 H(+) + O2 = 2 [Fe(III)-cytochrome b5] + an (11Z)-Delta(11)-fatty acyl-CoA + 2 H2O
(11Z)-hexadecenoyl-CoA + AH2 + O2 = (10E,12Z)-hexadecadienoyl-CoA + A + 2 H2O
(11Z)-hexadecenoyl-CoA + AH2 + O2 = (10E,12Z)-hexadecadienoyl-CoA + A + 2 H2O
Cofactor
Fe(2+)
Fe cation
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Keywords
Complete proteome
Fatty acid biosynthesis
Fatty acid metabolism
Iron
Lipid biosynthesis
Lipid metabolism
Membrane
Metal-binding
Oxidoreductase
Reference proteome
Transmembrane
Transmembrane helix
Endoplasmic reticulum
Feature
chain (11Z)-hexadec-11-enoyl-CoA conjugase
Uniprot
Q75PL7
Q9GU82
A0A1P1EST4
G8FQ78
Q4A181
D2SNV3
+ More
T1RU40 A0A2A4JG15 G8FQ79 A0A1V0M868 Q75PL5 A0A0U2F3F2 Q9NB26 A0A0U2F3E1 Q8MZZ2 G8FQ76 Q6A4M5 Q2YHQ4 A0A291P0P5 Q95VE7 Q8ISS3 A0A068FKA4 G8FQ77 A0A076FRH6 A9YRX6 Q6US81 O44390 M4PTD0 A0A0K2GV16 A0A1L8D6M1 Q95UJ3 B7SB74 T1RTE1 U5KFQ4 H9JF74 A0A0N1PJV1 A8QVZ1 E3TMU8 S4WAY4 A0A212FJD3 E3UYX7 A0A075BRA0 E3UYX8 B6CBS5 A0A0K0N452 A0A0L7L2G4 W5QM97 Q8WPC0 Q7KEQ2 A0A0F7R1B4 A0A1L8D6B5 A0A0F7R6V4 Q2V0N6 A0A1L8D6L2 A0A0F7R1B9 B8Q500 Q95UU3 G8FQ82 G8FQ83 A0A0F7R6V7 D8L7A6 A0A0F7R2Q5 A4KWE7 D8UUU1 D8L626 C0STP4 A4KWE9 A0A1L8D6C0 A0A194Q9S9 A0A0E3VMW5 A0A023EPC8 G8FQ80 A0A182MQV9 D8UUT8 Q17G31 A0A182VZL7 A0A182Y3S6 B0X4F1 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 A0A2M3YY30 A0A182K5W6 G8FQ74 A0A182PSJ2 W5J215 A0A182QY62 D6X4Z9 T1DNF2 A0A2M3YY13 A0A182RZM3 A0A1Q3FJ73 G8FQ73 A0A182IX38 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 A0A182NRU1
T1RU40 A0A2A4JG15 G8FQ79 A0A1V0M868 Q75PL5 A0A0U2F3F2 Q9NB26 A0A0U2F3E1 Q8MZZ2 G8FQ76 Q6A4M5 Q2YHQ4 A0A291P0P5 Q95VE7 Q8ISS3 A0A068FKA4 G8FQ77 A0A076FRH6 A9YRX6 Q6US81 O44390 M4PTD0 A0A0K2GV16 A0A1L8D6M1 Q95UJ3 B7SB74 T1RTE1 U5KFQ4 H9JF74 A0A0N1PJV1 A8QVZ1 E3TMU8 S4WAY4 A0A212FJD3 E3UYX7 A0A075BRA0 E3UYX8 B6CBS5 A0A0K0N452 A0A0L7L2G4 W5QM97 Q8WPC0 Q7KEQ2 A0A0F7R1B4 A0A1L8D6B5 A0A0F7R6V4 Q2V0N6 A0A1L8D6L2 A0A0F7R1B9 B8Q500 Q95UU3 G8FQ82 G8FQ83 A0A0F7R6V7 D8L7A6 A0A0F7R2Q5 A4KWE7 D8UUU1 D8L626 C0STP4 A4KWE9 A0A1L8D6C0 A0A194Q9S9 A0A0E3VMW5 A0A023EPC8 G8FQ80 A0A182MQV9 D8UUT8 Q17G31 A0A182VZL7 A0A182Y3S6 B0X4F1 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 A0A2M3YY30 A0A182K5W6 G8FQ74 A0A182PSJ2 W5J215 A0A182QY62 D6X4Z9 T1DNF2 A0A2M3YY13 A0A182RZM3 A0A1Q3FJ73 G8FQ73 A0A182IX38 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 A0A182NRU1
EC Number
1.14.19.15
1.14.19.5
1.14.19.5
Pubmed
10767556
15173596
19121390
22291612
17517337
20074338
+ More
24500170 28349297 28986331 11483431 12524345 12770579 15527989 26417103 15455060 26385554 24817326 18405835 25796477 15544945 9860961 11805319 20403437 24053512 26354079 17921252 20691782 24569486 22118469 21331247 18831750 26227816 24862548 12237399 16360168 19257880 12213232 17360547 21444802 24945155 17510324 25244985 20920257 23761445 18362917 20709164 19820115 20966253 12364791 14747013 17210077
24500170 28349297 28986331 11483431 12524345 12770579 15527989 26417103 15455060 26385554 24817326 18405835 25796477 15544945 9860961 11805319 20403437 24053512 26354079 17921252 20691782 24569486 22118469 21331247 18831750 26227816 24862548 12237399 16360168 19257880 12213232 17360547 21444802 24945155 17510324 25244985 20920257 23761445 18362917 20709164 19820115 20966253 12364791 14747013 17210077
EMBL
AF157627
AB166851
BABH01015400
AAF80355.1
BAD18122.1
AF182405
+ More
AAG16901.1 JN022488 AER29853.1 AM076339 CAJ27976.2 EZ407237 ACX53794.1 JX531654 AGO45839.1 NWSH01001511 PCG71017.1 JN022489 AER29854.1 KU755471 ARD71181.1 BABH01040457 AB166853 BAD18124.1 KP008124 MF687587 AKU76411.1 ATJ44513.1 AF272342 AAF81787.1 KP008117 AKU76404.1 AF482908 AAM28483.2 JN022486 AER29851.1 AF518014 AAQ12891.1 AM158251 CAJ43430.2 MF687528 ATJ44454.1 AF416738 AAL16642.1 AF545481 FJ999625 KJ622051 AID66659.1 JN022487 AER29852.1 KF960741 AII21943.1 EU285580 KM283200 ABX90049.1 AIU96339.1 AY362879 AF035375 AF297114 AF297113 KC422668 AGH12217.1 KP890026 ALA65425.1 GEYN01000135 JAV01994.1 AY049741 AAL11496.1 EU152400 FJ466458 ABX71809.1 ACS69071.1 JX531667 AGO45852.1 JX989153 AGR49312.1 BABH01026838 BABH01026839 BABH01026840 KQ460301 KPJ16138.1 EF150363 ABO43722.1 GU952763 ADO85596.1 JX964774 AGO96562.1 AGBW02008279 OWR53845.1 HM636633 ADP21587.1 JX679209 AGP26038.1 HM636634 ADP21588.1 EU152335 ABX71630.1 KM378664 AKJ32408.1 JTDY01003403 KOB69615.1 JQ978772 AGD98720.1 AF441861 AAL32060.1 AF441221 AAL35331.1 LC020021 BAR72981.1 GEYN01000136 JAV01993.1 LC020022 BAR72984.1 AB232855 BAE66602.1 GEYN01000122 JAV02007.1 LC020023 BAR72986.1 EU350083 EU350084 GQ166860 ACA81687.1 ACA81688.1 ADE97413.1 AY017379 AAG54077.1 JN022491 JN022492 AER29861.1 JN022493 AER29862.1 LC020024 BAR72989.1 FJ999626 ADC53486.1 BAR72990.1 EF113393 EF113394 ABO45226.1 ABO45227.1 GQ225753 ADE58526.1 FJ906800 ADB25212.1 AB465511 BAH56286.1 BAR72983.1 EF113395 EF113396 ABO45228.1 ABO45229.1 GEYN01000123 JAV02006.1 KQ459249 KPJ02273.1 AB910048 BAR45527.1 GAPW01002266 JAC11332.1 JN022490 AER29859.1 AXCM01000026 GQ225750 ADE58523.1 CH477266 EAT45554.1 EAT45555.1 DS232338 EDS40304.1 GGFK01006347 MBW39668.1 GGFJ01006787 MBW55928.1 GGFM01000428 MBW21179.1 JN022484 AER29858.1 ADMH02002143 ETN58292.1 AXCN02001148 HM234671 KQ971381 ADK13054.1 EEZ97602.1 GAMD01002940 JAA98650.1 GGFM01000421 MBW21172.1 GFDL01007467 JAV27578.1 JN022483 AER29857.1 AAAB01008987 EAA43119.4 EGK97500.1 EGK97501.1 EGK97502.1 APCN01000909
AAG16901.1 JN022488 AER29853.1 AM076339 CAJ27976.2 EZ407237 ACX53794.1 JX531654 AGO45839.1 NWSH01001511 PCG71017.1 JN022489 AER29854.1 KU755471 ARD71181.1 BABH01040457 AB166853 BAD18124.1 KP008124 MF687587 AKU76411.1 ATJ44513.1 AF272342 AAF81787.1 KP008117 AKU76404.1 AF482908 AAM28483.2 JN022486 AER29851.1 AF518014 AAQ12891.1 AM158251 CAJ43430.2 MF687528 ATJ44454.1 AF416738 AAL16642.1 AF545481 FJ999625 KJ622051 AID66659.1 JN022487 AER29852.1 KF960741 AII21943.1 EU285580 KM283200 ABX90049.1 AIU96339.1 AY362879 AF035375 AF297114 AF297113 KC422668 AGH12217.1 KP890026 ALA65425.1 GEYN01000135 JAV01994.1 AY049741 AAL11496.1 EU152400 FJ466458 ABX71809.1 ACS69071.1 JX531667 AGO45852.1 JX989153 AGR49312.1 BABH01026838 BABH01026839 BABH01026840 KQ460301 KPJ16138.1 EF150363 ABO43722.1 GU952763 ADO85596.1 JX964774 AGO96562.1 AGBW02008279 OWR53845.1 HM636633 ADP21587.1 JX679209 AGP26038.1 HM636634 ADP21588.1 EU152335 ABX71630.1 KM378664 AKJ32408.1 JTDY01003403 KOB69615.1 JQ978772 AGD98720.1 AF441861 AAL32060.1 AF441221 AAL35331.1 LC020021 BAR72981.1 GEYN01000136 JAV01993.1 LC020022 BAR72984.1 AB232855 BAE66602.1 GEYN01000122 JAV02007.1 LC020023 BAR72986.1 EU350083 EU350084 GQ166860 ACA81687.1 ACA81688.1 ADE97413.1 AY017379 AAG54077.1 JN022491 JN022492 AER29861.1 JN022493 AER29862.1 LC020024 BAR72989.1 FJ999626 ADC53486.1 BAR72990.1 EF113393 EF113394 ABO45226.1 ABO45227.1 GQ225753 ADE58526.1 FJ906800 ADB25212.1 AB465511 BAH56286.1 BAR72983.1 EF113395 EF113396 ABO45228.1 ABO45229.1 GEYN01000123 JAV02006.1 KQ459249 KPJ02273.1 AB910048 BAR45527.1 GAPW01002266 JAC11332.1 JN022490 AER29859.1 AXCM01000026 GQ225750 ADE58523.1 CH477266 EAT45554.1 EAT45555.1 DS232338 EDS40304.1 GGFK01006347 MBW39668.1 GGFJ01006787 MBW55928.1 GGFM01000428 MBW21179.1 JN022484 AER29858.1 ADMH02002143 ETN58292.1 AXCN02001148 HM234671 KQ971381 ADK13054.1 EEZ97602.1 GAMD01002940 JAA98650.1 GGFM01000421 MBW21172.1 GFDL01007467 JAV27578.1 JN022483 AER29857.1 AAAB01008987 EAA43119.4 EGK97500.1 EGK97501.1 EGK97502.1 APCN01000909
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000037510
UP000053268
+ More
UP000075883 UP000008820 UP000075920 UP000076408 UP000002320 UP000069272 UP000075881 UP000075885 UP000000673 UP000075886 UP000007266 UP000075900 UP000075880 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884
UP000075883 UP000008820 UP000075920 UP000076408 UP000002320 UP000069272 UP000075881 UP000075885 UP000000673 UP000075886 UP000007266 UP000075900 UP000075880 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884
Pfam
PF00487 FA_desaturase
ProteinModelPortal
Q75PL7
Q9GU82
A0A1P1EST4
G8FQ78
Q4A181
D2SNV3
+ More
T1RU40 A0A2A4JG15 G8FQ79 A0A1V0M868 Q75PL5 A0A0U2F3F2 Q9NB26 A0A0U2F3E1 Q8MZZ2 G8FQ76 Q6A4M5 Q2YHQ4 A0A291P0P5 Q95VE7 Q8ISS3 A0A068FKA4 G8FQ77 A0A076FRH6 A9YRX6 Q6US81 O44390 M4PTD0 A0A0K2GV16 A0A1L8D6M1 Q95UJ3 B7SB74 T1RTE1 U5KFQ4 H9JF74 A0A0N1PJV1 A8QVZ1 E3TMU8 S4WAY4 A0A212FJD3 E3UYX7 A0A075BRA0 E3UYX8 B6CBS5 A0A0K0N452 A0A0L7L2G4 W5QM97 Q8WPC0 Q7KEQ2 A0A0F7R1B4 A0A1L8D6B5 A0A0F7R6V4 Q2V0N6 A0A1L8D6L2 A0A0F7R1B9 B8Q500 Q95UU3 G8FQ82 G8FQ83 A0A0F7R6V7 D8L7A6 A0A0F7R2Q5 A4KWE7 D8UUU1 D8L626 C0STP4 A4KWE9 A0A1L8D6C0 A0A194Q9S9 A0A0E3VMW5 A0A023EPC8 G8FQ80 A0A182MQV9 D8UUT8 Q17G31 A0A182VZL7 A0A182Y3S6 B0X4F1 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 A0A2M3YY30 A0A182K5W6 G8FQ74 A0A182PSJ2 W5J215 A0A182QY62 D6X4Z9 T1DNF2 A0A2M3YY13 A0A182RZM3 A0A1Q3FJ73 G8FQ73 A0A182IX38 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 A0A182NRU1
T1RU40 A0A2A4JG15 G8FQ79 A0A1V0M868 Q75PL5 A0A0U2F3F2 Q9NB26 A0A0U2F3E1 Q8MZZ2 G8FQ76 Q6A4M5 Q2YHQ4 A0A291P0P5 Q95VE7 Q8ISS3 A0A068FKA4 G8FQ77 A0A076FRH6 A9YRX6 Q6US81 O44390 M4PTD0 A0A0K2GV16 A0A1L8D6M1 Q95UJ3 B7SB74 T1RTE1 U5KFQ4 H9JF74 A0A0N1PJV1 A8QVZ1 E3TMU8 S4WAY4 A0A212FJD3 E3UYX7 A0A075BRA0 E3UYX8 B6CBS5 A0A0K0N452 A0A0L7L2G4 W5QM97 Q8WPC0 Q7KEQ2 A0A0F7R1B4 A0A1L8D6B5 A0A0F7R6V4 Q2V0N6 A0A1L8D6L2 A0A0F7R1B9 B8Q500 Q95UU3 G8FQ82 G8FQ83 A0A0F7R6V7 D8L7A6 A0A0F7R2Q5 A4KWE7 D8UUU1 D8L626 C0STP4 A4KWE9 A0A1L8D6C0 A0A194Q9S9 A0A0E3VMW5 A0A023EPC8 G8FQ80 A0A182MQV9 D8UUT8 Q17G31 A0A182VZL7 A0A182Y3S6 B0X4F1 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 A0A2M3YY30 A0A182K5W6 G8FQ74 A0A182PSJ2 W5J215 A0A182QY62 D6X4Z9 T1DNF2 A0A2M3YY13 A0A182RZM3 A0A1Q3FJ73 G8FQ73 A0A182IX38 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 A0A182NRU1
PDB
4YMK
E-value=5.11683e-84,
Score=792
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Endoplasmic reticulum membrane
Endoplasmic reticulum membrane
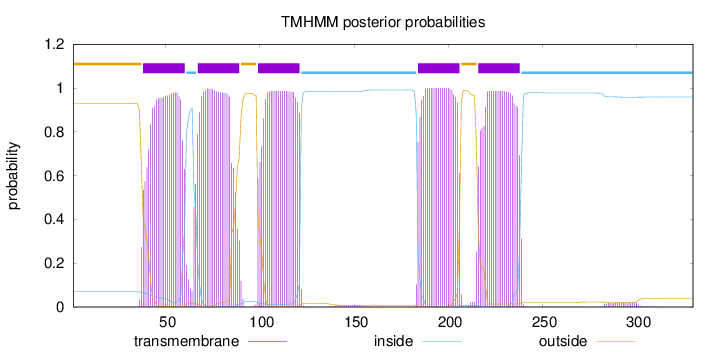
Length:
330
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
107.90838
Exp number, first 60 AAs:
20.32011
Total prob of N-in:
0.06941
POSSIBLE N-term signal
sequence
outside
1 - 37
TMhelix
38 - 60
inside
61 - 66
TMhelix
67 - 89
outside
90 - 98
TMhelix
99 - 121
inside
122 - 183
TMhelix
184 - 206
outside
207 - 215
TMhelix
216 - 238
inside
239 - 330
Population Genetic Test Statistics
Pi
189.59799
Theta
149.70327
Tajima's D
1.381563
CLR
0
CSRT
0.755512224388781
Interpretation
Uncertain
