Gene
KWMTBOMO13817
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_trace_amine-associated_receptor_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.708
Sequence
CDS
ATGTTCGACTCGACGACAAAAGGCGCATTGCTGGTAGCGCCTTCTCTATCAAACAGTTCGTGCTGGATGAATGGCAAAGGTTGGCCTTCGACTCTACCAGCACCCGGAGACATCGTAAAGGCTTTATTCATTTTTGCACTCTCCACTTCAATTGTCCTTGGGAATCTCATCTTAATCTGCGTTATGAATAATCGACGTTACATCAAATATATTGGAAATCAGCCAAGATACCTGCTGACGTCACTCGCTCTGAACGATTTGTTCACCGGAATCTTGATAGCTCCAGTGGCTCTTCTGCCTGCCCTTTACAAGTGTTGGCCGTATGGCGAGATATTTTGTCAGATACAGGCTCTGTTGAGGGGAGCCGTGAGTCAACAAAACGCCGTTATTCTCGTCTGCATGGCGATTGACCGTTACCTTTATTTGTTACACCCGAATACTTATCATAAGCATTCCAGCAAAAAGGTGAGATTCTTATGA
Protein
MFDSTTKGALLVAPSLSNSSCWMNGKGWPSTLPAPGDIVKALFIFALSTSIVLGNLILICVMNNRRYIKYIGNQPRYLLTSLALNDLFTGILIAPVALLPALYKCWPYGEIFCQIQALLRGAVSQQNAVILVCMAIDRYLYLLHPNTYHKHSSKKVRFL
Summary
Uniprot
A0A2W1BSQ1
A0A2A4JNN1
A0A2H1V842
A0A194QMJ0
A0A0N1IN82
A0A3L8DYU8
+ More
E2AB52 A0A087ZU11 A0A2A3EJH9 A0A195FGW2 U4UQZ6 A0A182UZF6 A0A151I8Q5 Q17NZ0 A0A182RLK8 A0A182FLI2 A0A151JLT2 A0A158NY54 N1NV95 A0A151WHE7 B0XCW9 A0A195BS88 E9IFZ6 A0A139WMY1 A0A2J7QCM0 A0A0L7QP35 A0A2P8YDK6 Q5TW58 A0A232F0U3 A0A1S4EW38 A0A182Y6Y9 A0A1J1IZZ7 Q9W167 B4IHH0 A0A1W4WD03 A0A0M3QV70 B4H669 A0A0B4KG01 B3NQK1 B4QBZ8 A0A0J9RKD9 A0A3B0J2N9 B3MFS4 Q28WW1 B4J532 B4MP22 B4LKF0 B4KU94 A0A182QWR3 B4PBA5 A0A1A9XVY1 A0A336LML6 A0A1S4ET98 A0A2H8TIR6 A0A1I8NDV5 A0A1B0AP18 A0A1I8NMT7 J9K798 A0A2S2N9I4 A0A2H8TY23 A0A0L7KQX1 A0A026W1T7 A0A154P9A1 F4W9K8 A0A1A9Z7M8 T1GQF2 A0A0K2T3B4 A0A1B0FQF0 A0A1A9WXJ0 A0A2S2QQ03 A0A336LJ54 V3ZA87 A0A0N7ZLV3 A0A0N8ADV0 A0A0P5ZBE1 A0A0N8EE56 A0A0P6IKL3 A0A0P5C8L6 A0A0P6BTX4 A0A0N8A9W8 A0A0P5AQG2 A0A0P6EXA3
E2AB52 A0A087ZU11 A0A2A3EJH9 A0A195FGW2 U4UQZ6 A0A182UZF6 A0A151I8Q5 Q17NZ0 A0A182RLK8 A0A182FLI2 A0A151JLT2 A0A158NY54 N1NV95 A0A151WHE7 B0XCW9 A0A195BS88 E9IFZ6 A0A139WMY1 A0A2J7QCM0 A0A0L7QP35 A0A2P8YDK6 Q5TW58 A0A232F0U3 A0A1S4EW38 A0A182Y6Y9 A0A1J1IZZ7 Q9W167 B4IHH0 A0A1W4WD03 A0A0M3QV70 B4H669 A0A0B4KG01 B3NQK1 B4QBZ8 A0A0J9RKD9 A0A3B0J2N9 B3MFS4 Q28WW1 B4J532 B4MP22 B4LKF0 B4KU94 A0A182QWR3 B4PBA5 A0A1A9XVY1 A0A336LML6 A0A1S4ET98 A0A2H8TIR6 A0A1I8NDV5 A0A1B0AP18 A0A1I8NMT7 J9K798 A0A2S2N9I4 A0A2H8TY23 A0A0L7KQX1 A0A026W1T7 A0A154P9A1 F4W9K8 A0A1A9Z7M8 T1GQF2 A0A0K2T3B4 A0A1B0FQF0 A0A1A9WXJ0 A0A2S2QQ03 A0A336LJ54 V3ZA87 A0A0N7ZLV3 A0A0N8ADV0 A0A0P5ZBE1 A0A0N8EE56 A0A0P6IKL3 A0A0P5C8L6 A0A0P6BTX4 A0A0N8A9W8 A0A0P5AQG2 A0A0P6EXA3
Pubmed
28756777
26354079
30249741
20798317
23537049
17510324
+ More
21347285 18054377 18025266 18316733 20068045 21843505 23604020 21282665 18362917 19820115 29403074 12364791 14747013 17210077 28648823 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 22936249 15632085 17550304 25315136 26227816 24508170 21719571 23254933
21347285 18054377 18025266 18316733 20068045 21843505 23604020 21282665 18362917 19820115 29403074 12364791 14747013 17210077 28648823 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 22936249 15632085 17550304 25315136 26227816 24508170 21719571 23254933
EMBL
KZ149950
PZC76674.1
NWSH01000992
PCG73194.1
ODYU01000984
SOQ36572.1
+ More
KQ461195 KPJ06747.1 KQ459037 KPJ04308.1 QOIP01000002 RLU25462.1 GL438234 EFN69328.1 KZ288227 PBC31858.1 KQ981606 KYN39477.1 KB632401 ERL94923.1 KQ978339 KYM95020.1 CH477195 EAT48408.2 KQ978986 KYN26725.1 ADTU01000555 ADTU01000556 ADTU01000557 ADTU01000558 BK005874 DAA64511.1 KQ983128 KYQ47225.1 DS232723 EDS45164.1 KQ976423 KYM88880.1 GL762905 EFZ20502.1 KQ971312 KYB29340.1 NEVH01016289 PNF26315.1 KQ414845 KOC60261.1 PYGN01000678 PSN42341.1 AAAB01008816 EAL41534.3 NNAY01001309 OXU24406.1 CVRI01000065 CRL05781.1 AE013599 AAF47210.2 CH480838 EDW49194.1 CP012524 ALC41899.1 CH479213 EDW33293.1 AGB93699.1 CH954179 EDV57004.2 CM000362 EDX08553.1 CM002911 KMY96336.1 OUUW01000001 SPP73363.1 CH902619 EDV37764.2 CM000071 EAL26555.3 CH916367 EDW00658.1 CH963848 EDW73861.2 CH940648 EDW60671.2 CH933808 EDW09690.2 AXCN02001111 CM000158 EDW92537.2 UFQT01000020 SSX17879.1 GFXV01002218 MBW14023.1 JXJN01001112 JXJN01001113 JXJN01001114 JXJN01001115 ABLF02038332 ABLF02038339 ABLF02038341 ABLF02038345 GGMR01001208 MBY13827.1 GFXV01007005 MBW18810.1 JTDY01006817 KOB65672.1 KK107555 EZA49019.1 KQ434849 KZC08485.1 GL888033 EGI68994.1 CAQQ02016704 HACA01002726 CDW20087.1 CCAG010017087 GGMS01010604 MBY79807.1 SSX17880.1 KB202823 ESO87863.1 GDIP01232713 JAI90688.1 GDIP01157137 JAJ66265.1 GDIP01046312 JAM57403.1 GDIQ01035620 JAN59117.1 GDIQ01009530 JAN85207.1 GDIP01192472 GDIP01190122 LRGB01000915 JAJ33280.1 KZS15243.1 GDIP01009573 JAM94142.1 GDIP01168193 JAJ55209.1 GDIP01195514 JAJ27888.1 GDIQ01069261 JAN25476.1
KQ461195 KPJ06747.1 KQ459037 KPJ04308.1 QOIP01000002 RLU25462.1 GL438234 EFN69328.1 KZ288227 PBC31858.1 KQ981606 KYN39477.1 KB632401 ERL94923.1 KQ978339 KYM95020.1 CH477195 EAT48408.2 KQ978986 KYN26725.1 ADTU01000555 ADTU01000556 ADTU01000557 ADTU01000558 BK005874 DAA64511.1 KQ983128 KYQ47225.1 DS232723 EDS45164.1 KQ976423 KYM88880.1 GL762905 EFZ20502.1 KQ971312 KYB29340.1 NEVH01016289 PNF26315.1 KQ414845 KOC60261.1 PYGN01000678 PSN42341.1 AAAB01008816 EAL41534.3 NNAY01001309 OXU24406.1 CVRI01000065 CRL05781.1 AE013599 AAF47210.2 CH480838 EDW49194.1 CP012524 ALC41899.1 CH479213 EDW33293.1 AGB93699.1 CH954179 EDV57004.2 CM000362 EDX08553.1 CM002911 KMY96336.1 OUUW01000001 SPP73363.1 CH902619 EDV37764.2 CM000071 EAL26555.3 CH916367 EDW00658.1 CH963848 EDW73861.2 CH940648 EDW60671.2 CH933808 EDW09690.2 AXCN02001111 CM000158 EDW92537.2 UFQT01000020 SSX17879.1 GFXV01002218 MBW14023.1 JXJN01001112 JXJN01001113 JXJN01001114 JXJN01001115 ABLF02038332 ABLF02038339 ABLF02038341 ABLF02038345 GGMR01001208 MBY13827.1 GFXV01007005 MBW18810.1 JTDY01006817 KOB65672.1 KK107555 EZA49019.1 KQ434849 KZC08485.1 GL888033 EGI68994.1 CAQQ02016704 HACA01002726 CDW20087.1 CCAG010017087 GGMS01010604 MBY79807.1 SSX17880.1 KB202823 ESO87863.1 GDIP01232713 JAI90688.1 GDIP01157137 JAJ66265.1 GDIP01046312 JAM57403.1 GDIQ01035620 JAN59117.1 GDIQ01009530 JAN85207.1 GDIP01192472 GDIP01190122 LRGB01000915 JAJ33280.1 KZS15243.1 GDIP01009573 JAM94142.1 GDIP01168193 JAJ55209.1 GDIP01195514 JAJ27888.1 GDIQ01069261 JAN25476.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000279307
UP000000311
UP000005203
+ More
UP000242457 UP000078541 UP000030742 UP000075903 UP000078542 UP000008820 UP000075900 UP000069272 UP000078492 UP000005205 UP000075809 UP000002320 UP000078540 UP000007266 UP000235965 UP000053825 UP000245037 UP000007062 UP000215335 UP000076408 UP000183832 UP000000803 UP000001292 UP000192221 UP000092553 UP000008744 UP000008711 UP000000304 UP000268350 UP000007801 UP000001819 UP000001070 UP000007798 UP000008792 UP000009192 UP000075886 UP000002282 UP000092443 UP000079169 UP000095301 UP000092460 UP000095300 UP000007819 UP000037510 UP000053097 UP000076502 UP000007755 UP000092445 UP000015102 UP000092444 UP000091820 UP000030746 UP000076858
UP000242457 UP000078541 UP000030742 UP000075903 UP000078542 UP000008820 UP000075900 UP000069272 UP000078492 UP000005205 UP000075809 UP000002320 UP000078540 UP000007266 UP000235965 UP000053825 UP000245037 UP000007062 UP000215335 UP000076408 UP000183832 UP000000803 UP000001292 UP000192221 UP000092553 UP000008744 UP000008711 UP000000304 UP000268350 UP000007801 UP000001819 UP000001070 UP000007798 UP000008792 UP000009192 UP000075886 UP000002282 UP000092443 UP000079169 UP000095301 UP000092460 UP000095300 UP000007819 UP000037510 UP000053097 UP000076502 UP000007755 UP000092445 UP000015102 UP000092444 UP000091820 UP000030746 UP000076858
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BSQ1
A0A2A4JNN1
A0A2H1V842
A0A194QMJ0
A0A0N1IN82
A0A3L8DYU8
+ More
E2AB52 A0A087ZU11 A0A2A3EJH9 A0A195FGW2 U4UQZ6 A0A182UZF6 A0A151I8Q5 Q17NZ0 A0A182RLK8 A0A182FLI2 A0A151JLT2 A0A158NY54 N1NV95 A0A151WHE7 B0XCW9 A0A195BS88 E9IFZ6 A0A139WMY1 A0A2J7QCM0 A0A0L7QP35 A0A2P8YDK6 Q5TW58 A0A232F0U3 A0A1S4EW38 A0A182Y6Y9 A0A1J1IZZ7 Q9W167 B4IHH0 A0A1W4WD03 A0A0M3QV70 B4H669 A0A0B4KG01 B3NQK1 B4QBZ8 A0A0J9RKD9 A0A3B0J2N9 B3MFS4 Q28WW1 B4J532 B4MP22 B4LKF0 B4KU94 A0A182QWR3 B4PBA5 A0A1A9XVY1 A0A336LML6 A0A1S4ET98 A0A2H8TIR6 A0A1I8NDV5 A0A1B0AP18 A0A1I8NMT7 J9K798 A0A2S2N9I4 A0A2H8TY23 A0A0L7KQX1 A0A026W1T7 A0A154P9A1 F4W9K8 A0A1A9Z7M8 T1GQF2 A0A0K2T3B4 A0A1B0FQF0 A0A1A9WXJ0 A0A2S2QQ03 A0A336LJ54 V3ZA87 A0A0N7ZLV3 A0A0N8ADV0 A0A0P5ZBE1 A0A0N8EE56 A0A0P6IKL3 A0A0P5C8L6 A0A0P6BTX4 A0A0N8A9W8 A0A0P5AQG2 A0A0P6EXA3
E2AB52 A0A087ZU11 A0A2A3EJH9 A0A195FGW2 U4UQZ6 A0A182UZF6 A0A151I8Q5 Q17NZ0 A0A182RLK8 A0A182FLI2 A0A151JLT2 A0A158NY54 N1NV95 A0A151WHE7 B0XCW9 A0A195BS88 E9IFZ6 A0A139WMY1 A0A2J7QCM0 A0A0L7QP35 A0A2P8YDK6 Q5TW58 A0A232F0U3 A0A1S4EW38 A0A182Y6Y9 A0A1J1IZZ7 Q9W167 B4IHH0 A0A1W4WD03 A0A0M3QV70 B4H669 A0A0B4KG01 B3NQK1 B4QBZ8 A0A0J9RKD9 A0A3B0J2N9 B3MFS4 Q28WW1 B4J532 B4MP22 B4LKF0 B4KU94 A0A182QWR3 B4PBA5 A0A1A9XVY1 A0A336LML6 A0A1S4ET98 A0A2H8TIR6 A0A1I8NDV5 A0A1B0AP18 A0A1I8NMT7 J9K798 A0A2S2N9I4 A0A2H8TY23 A0A0L7KQX1 A0A026W1T7 A0A154P9A1 F4W9K8 A0A1A9Z7M8 T1GQF2 A0A0K2T3B4 A0A1B0FQF0 A0A1A9WXJ0 A0A2S2QQ03 A0A336LJ54 V3ZA87 A0A0N7ZLV3 A0A0N8ADV0 A0A0P5ZBE1 A0A0N8EE56 A0A0P6IKL3 A0A0P5C8L6 A0A0P6BTX4 A0A0N8A9W8 A0A0P5AQG2 A0A0P6EXA3
PDB
6ME8
E-value=9.95111e-08,
Score=129
Ontologies
GO
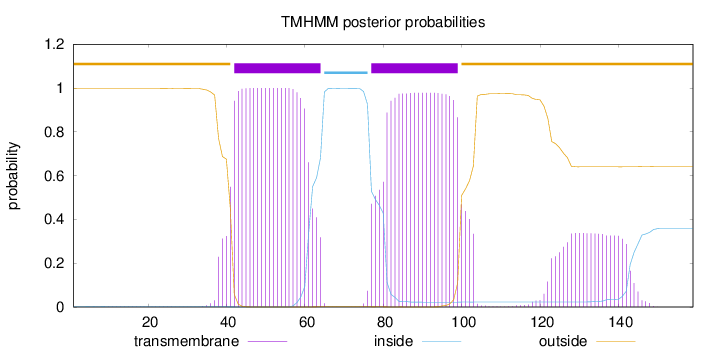
Topology
Length:
159
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
51.15487
Exp number, first 60 AAs:
20.26915
Total prob of N-in:
0.00313
POSSIBLE N-term signal
sequence
outside
1 - 41
TMhelix
42 - 64
inside
65 - 76
TMhelix
77 - 99
outside
100 - 159
Population Genetic Test Statistics
Pi
267.523229
Theta
171.78263
Tajima's D
1.539171
CLR
0.521868
CSRT
0.791310434478276
Interpretation
Uncertain
