Gene
KWMTBOMO13814
Pre Gene Modal
BGIBMGA011551
Annotation
PREDICTED:_beta-secretase_1-like_[Bombyx_mori]
Full name
Beta-secretase 1
Alternative Name
Beta-site amyloid precursor protein cleaving enzyme 1
Memapsin-2
Membrane-associated aspartic protease 2
Memapsin-2
Membrane-associated aspartic protease 2
Location in the cell
Extracellular Reliability : 1.726 PlasmaMembrane Reliability : 1.508
Sequence
CDS
ATGGCGAAGCTGTTAATAATTTTTCTCTATTGTATTTGTGCGAATGGAGAACAGTTTAATTTGCAAGGAGATGCCGGCCTTGCCTATTCTGTTGAGGTAATCATGGGACATCCACATCAAAAGATAAACCTTTTAATTGATACTGGTAGCACAACATTAGCTGTTGCTTCATATTCAAGACAAGATAGTGATAAATACTTTGATGCAAACAATTCTAGCAGCATTTATGACAGTGGGAAAAAGGTGCAGGCAACATACTTTGAAGGCAAATGGATTGGCCGTTTAGTCTCAGATTATATACAGCTCCCATCTATGCCATTGGTGCCAGAAGTCAGGACTGATATGGCACTTATTATGGAAAGCCACAATTTTTTTATGAATGGCTCACAATGGCAGGGTCTTCTAGGACTCGCATATCTACCAGTTGGAGCAAGGGGAGCAGATGTAATTGTGGAATCATGGTTAGATTCCCTTGACCGTACTTTGACTAGACCTGTATCATTTCAACTCAAACTTTGTGCTACATTAAGTACACAAAATGCGACGCACTATGGAAACTTTCAGATACTAGATGGTAAACAAAAAAATGAAAATTTAAACAAATCATACCGAACAACTATAATACAAAAAAGGTGGTATGAAGTTGGTGTTTTATCTGTGAGGGTTATGAGACATGTAAATAAATCAAGCAATCCTCCAGACATAGATGAACATATGTGTCAAACGCTTAATAAAATAAAGGCTATAGTTGATAGCGGAACAACAAATATAAGGTTACCTAATTGTTATTTTAGGCAGATTGTTGATGATTTGAGAAGCGCAGCACAGACTTCTAATATGTTAATTACGGATGACTTTTGGTATGCGGGAGAGACTGGATGTTGGCCAGAGCCTCAAGACTGGTCACTACCCTGGGTAGCGGTAGACCTACTTAGCTACGAAGCAGATAACCAGTACTTTACGTTAATGCTGTCGCCGCAGAATTATATGAGAGTCGTGGCAGCACAGAACAGCAGCGGTGAAAGTGGGACCGTATCAGAATACTGTTACAAATTGGGTTTTGAAAAAGGCGGAGATGAAACTGTACTCGGCTATACAGCCATGGAAGGATTTCAGATCAGCCGGTTGGATCGGCTGGCAGACCTCGGACTGTGGTCCAAACACTAA
Protein
MAKLLIIFLYCICANGEQFNLQGDAGLAYSVEVIMGHPHQKINLLIDTGSTTLAVASYSRQDSDKYFDANNSSSIYDSGKKVQATYFEGKWIGRLVSDYIQLPSMPLVPEVRTDMALIMESHNFFMNGSQWQGLLGLAYLPVGARGADVIVESWLDSLDRTLTRPVSFQLKLCATLSTQNATHYGNFQILDGKQKNENLNKSYRTTIIQKRWYEVGVLSVRVMRHVNKSSNPPDIDEHMCQTLNKIKAIVDSGTTNIRLPNCYFRQIVDDLRSAAQTSNMLITDDFWYAGETGCWPEPQDWSLPWVAVDLLSYEADNQYFTLMLSPQNYMRVVAAQNSSGESGTVSEYCYKLGFEKGGDETVLGYTAMEGFQISRLDRLADLGLWSKH
Summary
Description
Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase. Cleaves APP with much more catalytic efficiency than for the wild-type.
Catalytic Activity
Broad endopeptidase specificity. Cleaves Glu-Val-Asn-Leu-|-Asp-Ala-Glu-Phe in the Swedish variant of Alzheimer's amyloid precursor protein.
Subunit
Monomer. Interacts (via DXXLL motif) with GGA1, GGA2 and GGA3 (via their VHS domain); the interaction highly increases when BACE1 is phosphorylated at Ser-498. Interacts with RTN3 and RTN4. Interacts with SNX6. Interacts with PCSK9. Interacts with NAT8 and NAT8B. Interacts with BIN1 (By similarity).
Similarity
Belongs to the peptidase A1 family.
Keywords
Acetylation
Aspartyl protease
Cell membrane
Complete proteome
Cytoplasmic vesicle
Disulfide bond
Endoplasmic reticulum
Endosome
Glycoprotein
Golgi apparatus
Hydrolase
Isopeptide bond
Lipoprotein
Lysosome
Membrane
Palmitate
Phosphoprotein
Protease
Reference proteome
Signal
Transmembrane
Transmembrane helix
Ubl conjugation
Zymogen
Feature
chain Beta-secretase 1
Uniprot
A0A0N0P9Z3
A0A194QP43
A0A2W1BNY8
A0A2H1VDQ6
A0A3S2NRM5
A0A2A4J4D8
+ More
A0A1E1WAL9 A0A2P8XBR8 A0A2J7QRA6 A0A067QVR7 A0A2J7QRA2 A0A3L8S139 U3JTE5 W5MGP0 A0A3M0IYY5 A0A218UQP8 H2T442 A0A3Q2D4W4 H3DA27 A0A0Q3T1R0 F1N916 A0A226PMM9 A0A1V4J835 A0A3B3S824 A0A3N0Z3Y5 A0A1A8PDI3 A0A1A8I891 A0A1A8CH12 A0A1A8U632 A0A3P9NNE7 A0A1A8PRS4 A0A1A8G7W0 A0A3B3VJ97 A0A1A8MPD5 A0A3P9NNH4 A0A087XNU3 A0A3B3X3P6 A0A3B5QZG1 A0A0F8AXU5 A0A3B4BPE8 A0A2I4CNB6 A0A3B3BIR4 A0A151MVW4 G3NRK5 A0A3P8WBP3 A0A3Q0RU93 A0A3B1IHI0 A0A3B4X2Z0 H3BFZ7 A0A3Q0S6U0 A0A098LWN5 A0A1U7S779 I3KIR1 A0A3B5ATN3 A0A3Q1JPI7 A0A3Q3F7V2 A0A3P8XV70 A0A3P8PH74 A0A3P9BCK9 A0A3Q3BRF1 A0A3B4UH38 L5M0R1 A0A3Q3S981 A0A3B4UEA6 A0A3P9K9I4 A0A1W4YWI6 A0A3B4F5A5 A0A3Q3CER0 A0A060W7N0 K1PZA2 A0A2D0T8P4 G1PAB0 A0A3B3H7C2 A0A3P9HRM4 Q6NZT7 A0A3Q2QT07 A0A0A1WDW4 A0A1L8FFW8 A0A3Q1F609 A0A2U9B5T5 A0A3Q1LZR2 A0A1U7RGL7 A0A146W007 A0A098LX26 A0A3B4A1C3 Q8BQY4 A0A091DLE0 A0A287DDZ1 A0A2K6G6V5 A0A1S3NQS5 A0A1A6GT58 H0WUA0 V6F9A9 Q2HJ40 A0A1S3SYP0 A0A2K6G6W1 A0A060W0U5 A0A3P8U9P1
A0A1E1WAL9 A0A2P8XBR8 A0A2J7QRA6 A0A067QVR7 A0A2J7QRA2 A0A3L8S139 U3JTE5 W5MGP0 A0A3M0IYY5 A0A218UQP8 H2T442 A0A3Q2D4W4 H3DA27 A0A0Q3T1R0 F1N916 A0A226PMM9 A0A1V4J835 A0A3B3S824 A0A3N0Z3Y5 A0A1A8PDI3 A0A1A8I891 A0A1A8CH12 A0A1A8U632 A0A3P9NNE7 A0A1A8PRS4 A0A1A8G7W0 A0A3B3VJ97 A0A1A8MPD5 A0A3P9NNH4 A0A087XNU3 A0A3B3X3P6 A0A3B5QZG1 A0A0F8AXU5 A0A3B4BPE8 A0A2I4CNB6 A0A3B3BIR4 A0A151MVW4 G3NRK5 A0A3P8WBP3 A0A3Q0RU93 A0A3B1IHI0 A0A3B4X2Z0 H3BFZ7 A0A3Q0S6U0 A0A098LWN5 A0A1U7S779 I3KIR1 A0A3B5ATN3 A0A3Q1JPI7 A0A3Q3F7V2 A0A3P8XV70 A0A3P8PH74 A0A3P9BCK9 A0A3Q3BRF1 A0A3B4UH38 L5M0R1 A0A3Q3S981 A0A3B4UEA6 A0A3P9K9I4 A0A1W4YWI6 A0A3B4F5A5 A0A3Q3CER0 A0A060W7N0 K1PZA2 A0A2D0T8P4 G1PAB0 A0A3B3H7C2 A0A3P9HRM4 Q6NZT7 A0A3Q2QT07 A0A0A1WDW4 A0A1L8FFW8 A0A3Q1F609 A0A2U9B5T5 A0A3Q1LZR2 A0A1U7RGL7 A0A146W007 A0A098LX26 A0A3B4A1C3 Q8BQY4 A0A091DLE0 A0A287DDZ1 A0A2K6G6V5 A0A1S3NQS5 A0A1A6GT58 H0WUA0 V6F9A9 Q2HJ40 A0A1S3SYP0 A0A2K6G6W1 A0A060W0U5 A0A3P8U9P1
EC Number
3.4.23.46
Pubmed
26354079
28756777
29403074
24845553
30282656
21551351
+ More
15496914 15592404 24621616 29240929 23542700 25835551 29451363 22293439 24487278 25329095 9215903 25449103 25186727 25069045 17554307 24755649 22992520 21993624 27762356 19393038 28071753 25463417 10349636 11042159 11076861 11217851 12466851 16141073
15496914 15592404 24621616 29240929 23542700 25835551 29451363 22293439 24487278 25329095 9215903 25449103 25186727 25069045 17554307 24755649 22992520 21993624 27762356 19393038 28071753 25463417 10349636 11042159 11076861 11217851 12466851 16141073
EMBL
KQ459037
KPJ04306.1
KQ461195
KPJ06745.1
KZ149950
PZC76678.1
+ More
ODYU01001979 SOQ38921.1 RSAL01000004 RVE54625.1 NWSH01003295 PCG66596.1 GDQN01007004 JAT84050.1 PYGN01003650 PSN29455.1 NEVH01011917 PNF31121.1 KK853314 KDR08609.1 PNF31119.1 QUSF01000089 RLV93740.1 AGTO01001800 AHAT01018414 AHAT01018415 AHAT01018416 AHAT01018417 QRBI01000231 RMB91953.1 MUZQ01000188 OWK55692.1 LMAW01003036 KQK74518.1 AADN05000201 AWGT02000038 OXB81343.1 LSYS01008642 OPJ68289.1 RJVU01011952 ROL53209.1 HAEH01006380 SBR79077.1 HAED01006822 HAEE01010908 SBQ92948.1 HADZ01014130 HAEA01002783 SBP78071.1 HAEJ01002370 SBS42827.1 HAEI01003223 SBR83712.1 HAEB01019932 HAEC01000039 SBQ66459.1 HAEF01017408 HAEG01015172 SBR58567.1 AYCK01017144 KQ042328 KKF15961.1 AKHW03004818 KYO28703.1 AFYH01011934 AFYH01011935 AFYH01011936 GBHZ01000012 JAC94887.1 AERX01021523 KB105943 ELK31936.1 FR904426 CDQ63041.1 JH818774 EKC26973.1 AAPE02046303 AAPE02046304 AAPE02046305 AAPE02046306 AAPE02046307 AAPE02046308 BC065973 AAH65973.1 GBUG01000020 JAC96612.1 CM004479 OCT70472.1 CP026245 AWO99323.1 GCES01063437 JAR22886.1 GBIB01000010 JAC95042.1 AK046175 BAC32620.1 KN122326 KFO31095.1 AGTP01052904 LZPO01075906 OBS68517.1 AAQR03130901 AAQR03130902 GJ062397 DAA22394.1 BC113325 FR904313 CDQ58854.1
ODYU01001979 SOQ38921.1 RSAL01000004 RVE54625.1 NWSH01003295 PCG66596.1 GDQN01007004 JAT84050.1 PYGN01003650 PSN29455.1 NEVH01011917 PNF31121.1 KK853314 KDR08609.1 PNF31119.1 QUSF01000089 RLV93740.1 AGTO01001800 AHAT01018414 AHAT01018415 AHAT01018416 AHAT01018417 QRBI01000231 RMB91953.1 MUZQ01000188 OWK55692.1 LMAW01003036 KQK74518.1 AADN05000201 AWGT02000038 OXB81343.1 LSYS01008642 OPJ68289.1 RJVU01011952 ROL53209.1 HAEH01006380 SBR79077.1 HAED01006822 HAEE01010908 SBQ92948.1 HADZ01014130 HAEA01002783 SBP78071.1 HAEJ01002370 SBS42827.1 HAEI01003223 SBR83712.1 HAEB01019932 HAEC01000039 SBQ66459.1 HAEF01017408 HAEG01015172 SBR58567.1 AYCK01017144 KQ042328 KKF15961.1 AKHW03004818 KYO28703.1 AFYH01011934 AFYH01011935 AFYH01011936 GBHZ01000012 JAC94887.1 AERX01021523 KB105943 ELK31936.1 FR904426 CDQ63041.1 JH818774 EKC26973.1 AAPE02046303 AAPE02046304 AAPE02046305 AAPE02046306 AAPE02046307 AAPE02046308 BC065973 AAH65973.1 GBUG01000020 JAC96612.1 CM004479 OCT70472.1 CP026245 AWO99323.1 GCES01063437 JAR22886.1 GBIB01000010 JAC95042.1 AK046175 BAC32620.1 KN122326 KFO31095.1 AGTP01052904 LZPO01075906 OBS68517.1 AAQR03130901 AAQR03130902 GJ062397 DAA22394.1 BC113325 FR904313 CDQ58854.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000218220
UP000245037
UP000235965
+ More
UP000027135 UP000276834 UP000016665 UP000018468 UP000269221 UP000197619 UP000005226 UP000265020 UP000007303 UP000051836 UP000000539 UP000198419 UP000190648 UP000261540 UP000242638 UP000261500 UP000028760 UP000261480 UP000002852 UP000261440 UP000192220 UP000261560 UP000050525 UP000007635 UP000265120 UP000261340 UP000018467 UP000261360 UP000008672 UP000189705 UP000005207 UP000261400 UP000265040 UP000261660 UP000265140 UP000265100 UP000265160 UP000264800 UP000261420 UP000261640 UP000265180 UP000192224 UP000261460 UP000264840 UP000193380 UP000005408 UP000221080 UP000001074 UP000001038 UP000265200 UP000265000 UP000186698 UP000257200 UP000246464 UP000009136 UP000189706 UP000261520 UP000028990 UP000005215 UP000233160 UP000087266 UP000092124 UP000005225 UP000265080
UP000027135 UP000276834 UP000016665 UP000018468 UP000269221 UP000197619 UP000005226 UP000265020 UP000007303 UP000051836 UP000000539 UP000198419 UP000190648 UP000261540 UP000242638 UP000261500 UP000028760 UP000261480 UP000002852 UP000261440 UP000192220 UP000261560 UP000050525 UP000007635 UP000265120 UP000261340 UP000018467 UP000261360 UP000008672 UP000189705 UP000005207 UP000261400 UP000265040 UP000261660 UP000265140 UP000265100 UP000265160 UP000264800 UP000261420 UP000261640 UP000265180 UP000192224 UP000261460 UP000264840 UP000193380 UP000005408 UP000221080 UP000001074 UP000001038 UP000265200 UP000265000 UP000186698 UP000257200 UP000246464 UP000009136 UP000189706 UP000261520 UP000028990 UP000005215 UP000233160 UP000087266 UP000092124 UP000005225 UP000265080
PRIDE
Interpro
SUPFAM
SSF50630
SSF50630
Gene 3D
ProteinModelPortal
A0A0N0P9Z3
A0A194QP43
A0A2W1BNY8
A0A2H1VDQ6
A0A3S2NRM5
A0A2A4J4D8
+ More
A0A1E1WAL9 A0A2P8XBR8 A0A2J7QRA6 A0A067QVR7 A0A2J7QRA2 A0A3L8S139 U3JTE5 W5MGP0 A0A3M0IYY5 A0A218UQP8 H2T442 A0A3Q2D4W4 H3DA27 A0A0Q3T1R0 F1N916 A0A226PMM9 A0A1V4J835 A0A3B3S824 A0A3N0Z3Y5 A0A1A8PDI3 A0A1A8I891 A0A1A8CH12 A0A1A8U632 A0A3P9NNE7 A0A1A8PRS4 A0A1A8G7W0 A0A3B3VJ97 A0A1A8MPD5 A0A3P9NNH4 A0A087XNU3 A0A3B3X3P6 A0A3B5QZG1 A0A0F8AXU5 A0A3B4BPE8 A0A2I4CNB6 A0A3B3BIR4 A0A151MVW4 G3NRK5 A0A3P8WBP3 A0A3Q0RU93 A0A3B1IHI0 A0A3B4X2Z0 H3BFZ7 A0A3Q0S6U0 A0A098LWN5 A0A1U7S779 I3KIR1 A0A3B5ATN3 A0A3Q1JPI7 A0A3Q3F7V2 A0A3P8XV70 A0A3P8PH74 A0A3P9BCK9 A0A3Q3BRF1 A0A3B4UH38 L5M0R1 A0A3Q3S981 A0A3B4UEA6 A0A3P9K9I4 A0A1W4YWI6 A0A3B4F5A5 A0A3Q3CER0 A0A060W7N0 K1PZA2 A0A2D0T8P4 G1PAB0 A0A3B3H7C2 A0A3P9HRM4 Q6NZT7 A0A3Q2QT07 A0A0A1WDW4 A0A1L8FFW8 A0A3Q1F609 A0A2U9B5T5 A0A3Q1LZR2 A0A1U7RGL7 A0A146W007 A0A098LX26 A0A3B4A1C3 Q8BQY4 A0A091DLE0 A0A287DDZ1 A0A2K6G6V5 A0A1S3NQS5 A0A1A6GT58 H0WUA0 V6F9A9 Q2HJ40 A0A1S3SYP0 A0A2K6G6W1 A0A060W0U5 A0A3P8U9P1
A0A1E1WAL9 A0A2P8XBR8 A0A2J7QRA6 A0A067QVR7 A0A2J7QRA2 A0A3L8S139 U3JTE5 W5MGP0 A0A3M0IYY5 A0A218UQP8 H2T442 A0A3Q2D4W4 H3DA27 A0A0Q3T1R0 F1N916 A0A226PMM9 A0A1V4J835 A0A3B3S824 A0A3N0Z3Y5 A0A1A8PDI3 A0A1A8I891 A0A1A8CH12 A0A1A8U632 A0A3P9NNE7 A0A1A8PRS4 A0A1A8G7W0 A0A3B3VJ97 A0A1A8MPD5 A0A3P9NNH4 A0A087XNU3 A0A3B3X3P6 A0A3B5QZG1 A0A0F8AXU5 A0A3B4BPE8 A0A2I4CNB6 A0A3B3BIR4 A0A151MVW4 G3NRK5 A0A3P8WBP3 A0A3Q0RU93 A0A3B1IHI0 A0A3B4X2Z0 H3BFZ7 A0A3Q0S6U0 A0A098LWN5 A0A1U7S779 I3KIR1 A0A3B5ATN3 A0A3Q1JPI7 A0A3Q3F7V2 A0A3P8XV70 A0A3P8PH74 A0A3P9BCK9 A0A3Q3BRF1 A0A3B4UH38 L5M0R1 A0A3Q3S981 A0A3B4UEA6 A0A3P9K9I4 A0A1W4YWI6 A0A3B4F5A5 A0A3Q3CER0 A0A060W7N0 K1PZA2 A0A2D0T8P4 G1PAB0 A0A3B3H7C2 A0A3P9HRM4 Q6NZT7 A0A3Q2QT07 A0A0A1WDW4 A0A1L8FFW8 A0A3Q1F609 A0A2U9B5T5 A0A3Q1LZR2 A0A1U7RGL7 A0A146W007 A0A098LX26 A0A3B4A1C3 Q8BQY4 A0A091DLE0 A0A287DDZ1 A0A2K6G6V5 A0A1S3NQS5 A0A1A6GT58 H0WUA0 V6F9A9 Q2HJ40 A0A1S3SYP0 A0A2K6G6W1 A0A060W0U5 A0A3P8U9P1
PDB
6BFX
E-value=5.60752e-46,
Score=464
Ontologies
GO
GO:0016021
GO:0004190
GO:0005764
GO:0005769
GO:0050435
GO:0009986
GO:0030424
GO:0005802
GO:0055037
GO:2000300
GO:0005887
GO:0043525
GO:0001540
GO:0005771
GO:0098686
GO:0006509
GO:0019899
GO:0098793
GO:0005768
GO:0006508
GO:0005794
GO:0030163
GO:0016020
GO:0022011
GO:0031643
GO:0004175
GO:0005783
GO:0005770
GO:0045121
GO:0030659
GO:0016791
GO:0016075
GO:0003677
GO:0005524
GO:0016787
GO:0004713
GO:0016491
GO:0015930
PANTHER
Topology
Subcellular location
Cell membrane
Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Golgi apparatus Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
trans-Golgi network Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Endoplasmic reticulum Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Cell surface Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Cytoplasmic vesicle membrane Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Membrane raft Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Lysosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Late endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Early endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Recycling endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Golgi apparatus Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
trans-Golgi network Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Endoplasmic reticulum Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Cell surface Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Cytoplasmic vesicle membrane Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Membrane raft Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Lysosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Late endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Early endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
Recycling endosome Predominantly localized to the later Golgi/trans-Golgi network (TGN) and minimally detectable in the early Golgi compartments. A small portion is also found in the endoplasmic reticulum, endosomes and on the cell surface (By similarity). Colocalization with APP in early endosomes is due to addition of bisecting N-acetylglucosamine wich blocks targeting to late endosomes and lysosomes (By similarity). Retrogradly transported from endosomal compartments to the trans-Golgi network in a phosphorylation- and GGA1- dependent manner (By similarity). With evidence from 2 publications.
SignalP
Position: 1 - 16,
Likelihood: 0.977551
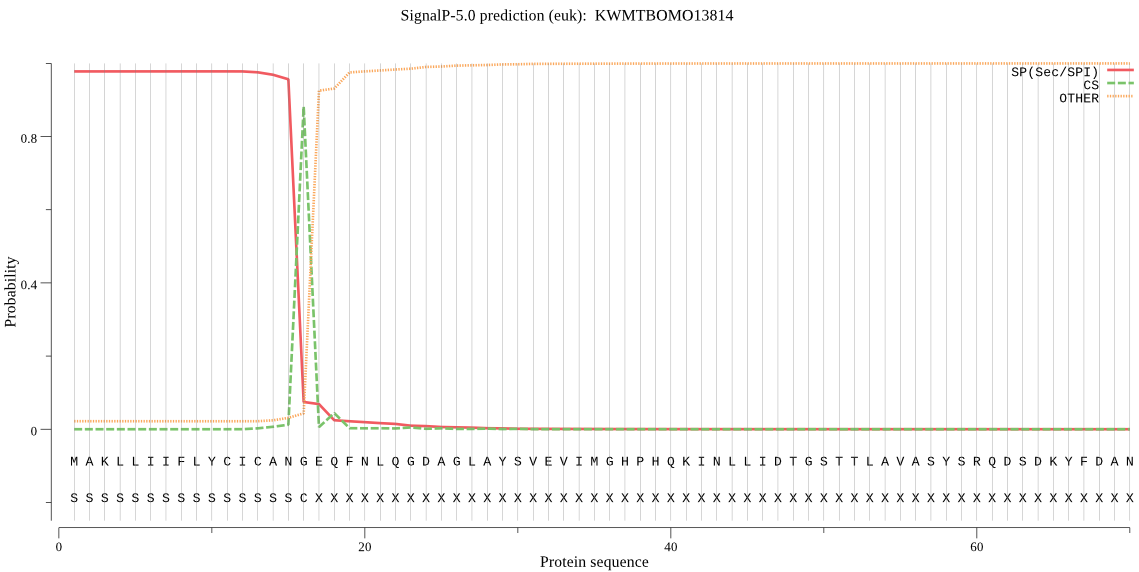
Length:
388
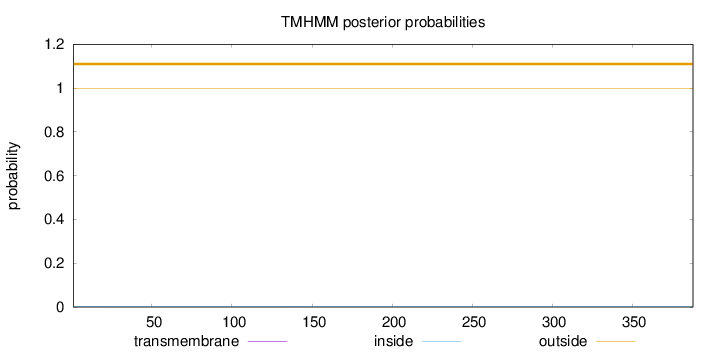
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01044
Exp number, first 60 AAs:
0.0063
Total prob of N-in:
0.00217
outside
1 - 388
Population Genetic Test Statistics
Pi
254.515329
Theta
190.445741
Tajima's D
1.106975
CLR
0.246682
CSRT
0.683965801709915
Interpretation
Uncertain
