Gene
KWMTBOMO13810
Pre Gene Modal
BGIBMGA011462
Annotation
PREDICTED:_alkaline_phosphatase-like_[Amyelois_transitella]
Full name
Alkaline phosphatase
Location in the cell
Mitochondrial Reliability : 1.619
Sequence
CDS
ATGAGTACTTGGTGTTGCATCGCGATCATGATGTCCACTATCAGTACCGGCGTCCTTTCACCAGCACCTCAGAAGACCGAAGTTCATGAGGCCTACGACCGCCAATATTGGTACACCCAAGCACAGGAAACATTACAGAAACGCCTTCAATACTCGACTGACAAAAAACCGGTAGCCAAAAACGTGATCCTTGTGGTTGGCGACGGTATGAGTCTTACAACAGCTACTGCCGCGAGGATACTTCGGGGGCAACGGAGGGGGCAATCTGGCGAAGATACTGATTTAGTGTGGGATACTTTCCCCTCGGTAGCTTTAGCTAAAACATACAACATAGACGCACAGACGGGCGAGTCGTCGGCTTGTGCGACAGCGCTGCTGTGCGGTGTGAAGGCGCGATATGAAACACTTGGATTAGATGCTGGCGGTCGTTTTAACAACTGCGCTTCAACTATACATTCAAAAGTGCCTTCTCTCGTTGACTGGGCCCATGAGGCTGGAAAATCAAGTGGGATAGTGACGACAGCGAGAGTAACACATGCGACACCGGCGGCGTTATATGCTCATGCCCCCTCACGTTACTGGGAGGATGACAGTCGAGTCCCTCCGACAGTTAGGAAGGACTGCAAGGATATTGCTCTGCAATTGGTCGAAAATGAACCAGGACGTAATATAAATGTAATTATGGGTGGTGGTAGAAGACACTTTCTACCAACAATCACAACGGATCCTGAACATCCGAACCGTGAAGGTCGAAGACTGGATGGCAGGAATCTCGCCGAAGACTGGGCTAGAGAAAAGAAAAGACGACGGTTGAGAGCTCAGTACATACATTCAAAGGAGCAATTGGCAAAACTGGATCCACGCACGGTGGATTATCTTTTAGGTCTCTTTGAATATTCTCATATGGAATTCAACGCAGAGCGCGGGGCGCTTGTTGACGCAGAAGAGGGAAGCGGAACGAGCACAACTGTGAAAGCCGACGACCCTTCTCTAGCCGACATGACTCGAGCCGCCCTTTCAATATTACTGAAAAATGACAAAGGATTTTTCTTGCTTATTGAAGGCGGACGTATAGACCACGCTCATCACTACAATAATCCGTACAGAGCTCTCGATGAAACGTTAGAGTTAGAAACCGCATTATTGGCTGCACTAGAGAGGGTAAATCCGGCGGAAACTTTGATCGTAGTAACCGCCGATCACGGTCACGTAATGACCTTTGGCGGTCAAGCAACGCCAAGAGGACATCCTGTCTTGGGTGCTGACACAGTGGTATCGGACATAGATGGACTACGCTACACGACTTTGTTGTACGGAACTGGACCTGGTCATTCTGAGCCTAGAGCGTTGCCGTTCAATTCAAGTAGCTCAACAAATCAAGCTGATGCGGTCCACGCTGCAGCTGTTCCAAGACAATGGGCTACGCATGGTGGCGAGGACGTACCAATATACGCATTAGGTCCGATGGCTACAATTTTATTTGCTGGAGTTGTAGAACAAAGCTACATACCTCATGCCATAGCATATGCAGCTTGTTTGGCGCATCACGCAAAGCGTTGTCAAGAAAAAATCAACTTTACGCAACCTTTAATTAAAGAAAAAGTAGCAAAATGCGTTCCGCCCGAAGTAAGTAGTGTTTCTGCGGCGGAAGACGTGAGAATCGACAATCCGAGTGGCCGTCGCATCGTGGTCGCGTCGAGCGTCATGTCCGACGAACGTGTACCGCGTTCTGCCGCTCCGCTTATCGCTACGCTTCTCACGGAACGCTACCTTATCTTAAGCATACTTAAATTGGTCGGATTCTTACGGATCGCTCGTTTTATCTTAGGCCTATAA
Protein
MSTWCCIAIMMSTISTGVLSPAPQKTEVHEAYDRQYWYTQAQETLQKRLQYSTDKKPVAKNVILVVGDGMSLTTATAARILRGQRRGQSGEDTDLVWDTFPSVALAKTYNIDAQTGESSACATALLCGVKARYETLGLDAGGRFNNCASTIHSKVPSLVDWAHEAGKSSGIVTTARVTHATPAALYAHAPSRYWEDDSRVPPTVRKDCKDIALQLVENEPGRNINVIMGGGRRHFLPTITTDPEHPNREGRRLDGRNLAEDWAREKKRRRLRAQYIHSKEQLAKLDPRTVDYLLGLFEYSHMEFNAERGALVDAEEGSGTSTTVKADDPSLADMTRAALSILLKNDKGFFLLIEGGRIDHAHHYNNPYRALDETLELETALLAALERVNPAETLIVVTADHGHVMTFGGQATPRGHPVLGADTVVSDIDGLRYTTLLYGTGPGHSEPRALPFNSSSSTNQADAVHAAAVPRQWATHGGEDVPIYALGPMATILFAGVVEQSYIPHAIAYAACLAHHAKRCQEKINFTQPLIKEKVAKCVPPEVSSVSAAEDVRIDNPSGRRIVVASSVMSDERVPRSAAPLIATLLTERYLILSILKLVGFLRIARFILGL
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Similarity
Belongs to the alkaline phosphatase family.
Feature
chain Alkaline phosphatase
Uniprot
A0A2A4ITJ3
A0A2W1BNN8
A0A212FJJ5
A0A0N1PG25
A0A194RPT5
A0A1Y1MXI2
+ More
A0A139WNY2 A0A1W4X780 A0A2L1IQ98 A0A1B6EBV1 A0A1B6LKY4 A0A067RT96 E0VJV3 A0A2L1IQ99 A0A2S2QCC1 A0A2H8TN21 A0A3L8DWC4 A0A026X2J0 A0A2J7PS94 A0A0C9QHH3 A0A088AKH3 A0A195F6L9 A0A151X769 A0A195CT65 A0A151IZ22 A0A087SWZ5 A0A195ATQ8 A0A0L7QZV5 A0A336LPP2 A0A2L2YN05 A0A087T3L1 A0A1S3JAP8 A0A1W7RB25 A0A0P7YCJ1 A0A1W4XZF9 Q32NT2 H9GA51 Q8AYA9 Q7ZYJ4 A0A1W4XPE6 W5M6U5 A0A087SZK6 A0A3Q3B3R2 A0A3Q3APK3 A0A2I4BBB2 A0A0P7Y253 A0A1W4ZZA1 F7EGD2 A0A3Q2WS28 A0A3Q3GIX9 A0A3B3SFK9 H3D3S4 Q4S777 A0A3L8RYU0 C3XYT0 A0A1L8FHH2 A0A287BSC3 A0A3P9AV06 A0A3P8NEJ0 A0A3B4F6X0 C3YZE2 A0A2I4BBE4 B5X1S3 A0A3Q3WR19 A0A3Q4GUG0 A0A3Q4BF09 A0A087SZK5 I3IW72 Q8AXY2 A0A2Y9LKT6 G3NL30 A0A0B6YV85 A0A3Q4BF13 A0A2G8LDM7 A0A383ZJU7 A0A3Q3IG72 U3JM59 A0A3B4CSJ8 A0A340XCS3 A0A2Y9EYY0 V9KSG4 A0A2Y9K958 A0A341D710 A0A3P8W4M6 A0A2Y9DN62 H2SDY1 A0A3P8XYH8 Q8AXY3 A0A3L7IAD7 H0X4F4 A0A3P8ZUF1 A0A151MJC1
A0A139WNY2 A0A1W4X780 A0A2L1IQ98 A0A1B6EBV1 A0A1B6LKY4 A0A067RT96 E0VJV3 A0A2L1IQ99 A0A2S2QCC1 A0A2H8TN21 A0A3L8DWC4 A0A026X2J0 A0A2J7PS94 A0A0C9QHH3 A0A088AKH3 A0A195F6L9 A0A151X769 A0A195CT65 A0A151IZ22 A0A087SWZ5 A0A195ATQ8 A0A0L7QZV5 A0A336LPP2 A0A2L2YN05 A0A087T3L1 A0A1S3JAP8 A0A1W7RB25 A0A0P7YCJ1 A0A1W4XZF9 Q32NT2 H9GA51 Q8AYA9 Q7ZYJ4 A0A1W4XPE6 W5M6U5 A0A087SZK6 A0A3Q3B3R2 A0A3Q3APK3 A0A2I4BBB2 A0A0P7Y253 A0A1W4ZZA1 F7EGD2 A0A3Q2WS28 A0A3Q3GIX9 A0A3B3SFK9 H3D3S4 Q4S777 A0A3L8RYU0 C3XYT0 A0A1L8FHH2 A0A287BSC3 A0A3P9AV06 A0A3P8NEJ0 A0A3B4F6X0 C3YZE2 A0A2I4BBE4 B5X1S3 A0A3Q3WR19 A0A3Q4GUG0 A0A3Q4BF09 A0A087SZK5 I3IW72 Q8AXY2 A0A2Y9LKT6 G3NL30 A0A0B6YV85 A0A3Q4BF13 A0A2G8LDM7 A0A383ZJU7 A0A3Q3IG72 U3JM59 A0A3B4CSJ8 A0A340XCS3 A0A2Y9EYY0 V9KSG4 A0A2Y9K958 A0A341D710 A0A3P8W4M6 A0A2Y9DN62 H2SDY1 A0A3P8XYH8 Q8AXY3 A0A3L7IAD7 H0X4F4 A0A3P8ZUF1 A0A151MJC1
EC Number
3.1.3.1
Pubmed
EMBL
NWSH01008033
PCG62718.1
KZ149950
PZC76682.1
AGBW02008260
OWR53902.1
+ More
KQ459037 KPJ04301.1 KQ459833 KPJ19868.1 GEZM01018032 JAV90373.1 KQ971311 KYB29475.1 MF741669 AVD96952.1 GEDC01001882 JAS35416.1 GEBQ01015629 JAT24348.1 KK852429 KDR24035.1 DS235230 EEB13659.1 MF741671 AVD96954.1 GGMS01006174 MBY75377.1 GFXV01003516 MBW15321.1 QOIP01000004 RLU24078.1 KK107024 EZA62298.1 NEVH01021937 PNF19205.1 GBYB01014088 JAG83855.1 KQ981756 KYN36108.1 KQ982450 KYQ56217.1 KQ977305 KYN03712.1 KQ980735 KYN13757.1 KK112338 KFM57384.1 KQ976741 KYM75581.1 KQ414672 KOC64154.1 UFQT01000099 SSX19880.1 IAAA01039428 LAA09377.1 KK113248 KFM59700.1 GFAH01000056 JAV48333.1 JARO02007446 KPP63948.1 BC108491 AAI08492.1 AAWZ02033950 AF539792 AAN31766.1 BC043760 CM004478 AAH43760.1 OCT73367.1 AHAT01009727 KK112695 KFM58295.1 JARO02011663 KPP59742.1 CAAE01014721 CAG03505.1 QUSF01000139 RLV89718.1 GG666473 EEN66965.1 CM004479 OCT71036.1 AEMK02000045 GG666566 EEN54213.1 BT044992 ACI33254.1 KFM58294.1 AERX01018584 AERX01018585 AERX01018586 AY145129 AAN64271.1 HACG01013127 HACG01013128 CEK59992.1 CEK59993.1 MRZV01000115 PIK58343.1 AGTO01022000 JW869282 AFP01800.1 AY144373 AAN46665.1 RAZU01000087 RLQ74541.1 AAQR03020624 AAQR03020625 AAQR03020626 AAQR03020627 AKHW03006061 KYO24632.1
KQ459037 KPJ04301.1 KQ459833 KPJ19868.1 GEZM01018032 JAV90373.1 KQ971311 KYB29475.1 MF741669 AVD96952.1 GEDC01001882 JAS35416.1 GEBQ01015629 JAT24348.1 KK852429 KDR24035.1 DS235230 EEB13659.1 MF741671 AVD96954.1 GGMS01006174 MBY75377.1 GFXV01003516 MBW15321.1 QOIP01000004 RLU24078.1 KK107024 EZA62298.1 NEVH01021937 PNF19205.1 GBYB01014088 JAG83855.1 KQ981756 KYN36108.1 KQ982450 KYQ56217.1 KQ977305 KYN03712.1 KQ980735 KYN13757.1 KK112338 KFM57384.1 KQ976741 KYM75581.1 KQ414672 KOC64154.1 UFQT01000099 SSX19880.1 IAAA01039428 LAA09377.1 KK113248 KFM59700.1 GFAH01000056 JAV48333.1 JARO02007446 KPP63948.1 BC108491 AAI08492.1 AAWZ02033950 AF539792 AAN31766.1 BC043760 CM004478 AAH43760.1 OCT73367.1 AHAT01009727 KK112695 KFM58295.1 JARO02011663 KPP59742.1 CAAE01014721 CAG03505.1 QUSF01000139 RLV89718.1 GG666473 EEN66965.1 CM004479 OCT71036.1 AEMK02000045 GG666566 EEN54213.1 BT044992 ACI33254.1 KFM58294.1 AERX01018584 AERX01018585 AERX01018586 AY145129 AAN64271.1 HACG01013127 HACG01013128 CEK59992.1 CEK59993.1 MRZV01000115 PIK58343.1 AGTO01022000 JW869282 AFP01800.1 AY144373 AAN46665.1 RAZU01000087 RLQ74541.1 AAQR03020624 AAQR03020625 AAQR03020626 AAQR03020627 AKHW03006061 KYO24632.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000007266
UP000192223
+ More
UP000027135 UP000009046 UP000279307 UP000053097 UP000235965 UP000005203 UP000078541 UP000075809 UP000078542 UP000078492 UP000054359 UP000078540 UP000053825 UP000085678 UP000034805 UP000192224 UP000001646 UP000186698 UP000018468 UP000264800 UP000192220 UP000002280 UP000264840 UP000261540 UP000007303 UP000276834 UP000001554 UP000008227 UP000265160 UP000265100 UP000261460 UP000087266 UP000261620 UP000261580 UP000005207 UP000248483 UP000007635 UP000230750 UP000261600 UP000016665 UP000261440 UP000265300 UP000248482 UP000252040 UP000265120 UP000248480 UP000005226 UP000265140 UP000273346 UP000005225 UP000050525
UP000027135 UP000009046 UP000279307 UP000053097 UP000235965 UP000005203 UP000078541 UP000075809 UP000078542 UP000078492 UP000054359 UP000078540 UP000053825 UP000085678 UP000034805 UP000192224 UP000001646 UP000186698 UP000018468 UP000264800 UP000192220 UP000002280 UP000264840 UP000261540 UP000007303 UP000276834 UP000001554 UP000008227 UP000265160 UP000265100 UP000261460 UP000087266 UP000261620 UP000261580 UP000005207 UP000248483 UP000007635 UP000230750 UP000261600 UP000016665 UP000261440 UP000265300 UP000248482 UP000252040 UP000265120 UP000248480 UP000005226 UP000265140 UP000273346 UP000005225 UP000050525
Pfam
PF00245 Alk_phosphatase
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
CDD
ProteinModelPortal
A0A2A4ITJ3
A0A2W1BNN8
A0A212FJJ5
A0A0N1PG25
A0A194RPT5
A0A1Y1MXI2
+ More
A0A139WNY2 A0A1W4X780 A0A2L1IQ98 A0A1B6EBV1 A0A1B6LKY4 A0A067RT96 E0VJV3 A0A2L1IQ99 A0A2S2QCC1 A0A2H8TN21 A0A3L8DWC4 A0A026X2J0 A0A2J7PS94 A0A0C9QHH3 A0A088AKH3 A0A195F6L9 A0A151X769 A0A195CT65 A0A151IZ22 A0A087SWZ5 A0A195ATQ8 A0A0L7QZV5 A0A336LPP2 A0A2L2YN05 A0A087T3L1 A0A1S3JAP8 A0A1W7RB25 A0A0P7YCJ1 A0A1W4XZF9 Q32NT2 H9GA51 Q8AYA9 Q7ZYJ4 A0A1W4XPE6 W5M6U5 A0A087SZK6 A0A3Q3B3R2 A0A3Q3APK3 A0A2I4BBB2 A0A0P7Y253 A0A1W4ZZA1 F7EGD2 A0A3Q2WS28 A0A3Q3GIX9 A0A3B3SFK9 H3D3S4 Q4S777 A0A3L8RYU0 C3XYT0 A0A1L8FHH2 A0A287BSC3 A0A3P9AV06 A0A3P8NEJ0 A0A3B4F6X0 C3YZE2 A0A2I4BBE4 B5X1S3 A0A3Q3WR19 A0A3Q4GUG0 A0A3Q4BF09 A0A087SZK5 I3IW72 Q8AXY2 A0A2Y9LKT6 G3NL30 A0A0B6YV85 A0A3Q4BF13 A0A2G8LDM7 A0A383ZJU7 A0A3Q3IG72 U3JM59 A0A3B4CSJ8 A0A340XCS3 A0A2Y9EYY0 V9KSG4 A0A2Y9K958 A0A341D710 A0A3P8W4M6 A0A2Y9DN62 H2SDY1 A0A3P8XYH8 Q8AXY3 A0A3L7IAD7 H0X4F4 A0A3P8ZUF1 A0A151MJC1
A0A139WNY2 A0A1W4X780 A0A2L1IQ98 A0A1B6EBV1 A0A1B6LKY4 A0A067RT96 E0VJV3 A0A2L1IQ99 A0A2S2QCC1 A0A2H8TN21 A0A3L8DWC4 A0A026X2J0 A0A2J7PS94 A0A0C9QHH3 A0A088AKH3 A0A195F6L9 A0A151X769 A0A195CT65 A0A151IZ22 A0A087SWZ5 A0A195ATQ8 A0A0L7QZV5 A0A336LPP2 A0A2L2YN05 A0A087T3L1 A0A1S3JAP8 A0A1W7RB25 A0A0P7YCJ1 A0A1W4XZF9 Q32NT2 H9GA51 Q8AYA9 Q7ZYJ4 A0A1W4XPE6 W5M6U5 A0A087SZK6 A0A3Q3B3R2 A0A3Q3APK3 A0A2I4BBB2 A0A0P7Y253 A0A1W4ZZA1 F7EGD2 A0A3Q2WS28 A0A3Q3GIX9 A0A3B3SFK9 H3D3S4 Q4S777 A0A3L8RYU0 C3XYT0 A0A1L8FHH2 A0A287BSC3 A0A3P9AV06 A0A3P8NEJ0 A0A3B4F6X0 C3YZE2 A0A2I4BBE4 B5X1S3 A0A3Q3WR19 A0A3Q4GUG0 A0A3Q4BF09 A0A087SZK5 I3IW72 Q8AXY2 A0A2Y9LKT6 G3NL30 A0A0B6YV85 A0A3Q4BF13 A0A2G8LDM7 A0A383ZJU7 A0A3Q3IG72 U3JM59 A0A3B4CSJ8 A0A340XCS3 A0A2Y9EYY0 V9KSG4 A0A2Y9K958 A0A341D710 A0A3P8W4M6 A0A2Y9DN62 H2SDY1 A0A3P8XYH8 Q8AXY3 A0A3L7IAD7 H0X4F4 A0A3P8ZUF1 A0A151MJC1
PDB
4KJG
E-value=1.08935e-91,
Score=861
Ontologies
PATHWAY
GO
PANTHER
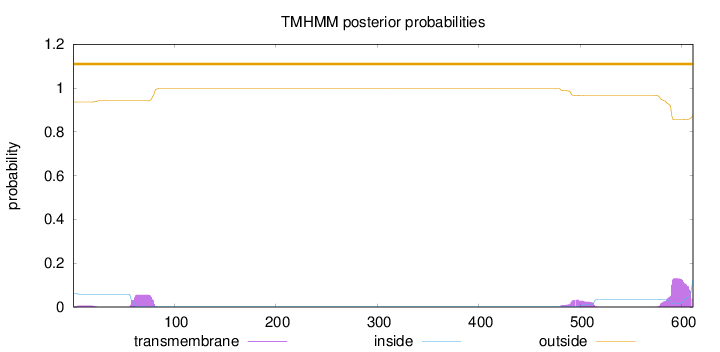
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.822129
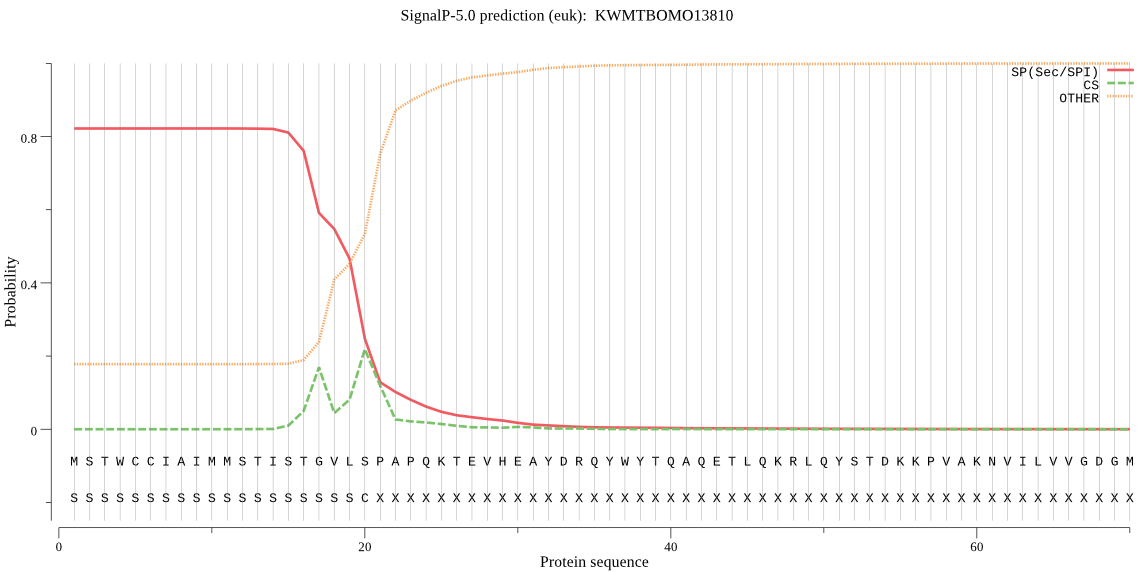
Length:
611
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.68756
Exp number, first 60 AAs:
0.22004
Total prob of N-in:
0.06398
outside
1 - 611
Population Genetic Test Statistics
Pi
280.95677
Theta
176.419454
Tajima's D
1.353971
CLR
0.395224
CSRT
0.759062046897655
Interpretation
Uncertain
