Gene
KWMTBOMO13804
Annotation
PREDICTED:_ubiquinone_biosynthesis_O-methyltransferase?_mitochondrial-like_[Bombyx_mori]
Full name
Ubiquinone biosynthesis O-methyltransferase, mitochondrial
Alternative Name
3-demethylubiquinol 3-O-methyltransferase
Polyprenyldihydroxybenzoate methyltransferase
Polyprenyldihydroxybenzoate methyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.46
Sequence
CDS
ATGCGAAAAATATTTTTCCATTTAATTTTGGTAACAGTTATCGTTTTTTGGACACTAATAAAGGTCCCAAAGCTCCTGCATCAGTACCTCTCAGAAAGAAGAGTGACACACGAACGTAAAGTAACAGAATGCGATGAAAATTCAACAGTAATAGAACAAAACGTCGAAATTTTTGCAAATCTAGATGATTGGTGGAATCCTCGTGGAAAACTTTCTACATTACACAAATTCAACGTTATAAGAATTCCGTTCATTCAAAATGGTATACTTGCTCACACCGGAATAAACACTCTGAATAATATAAAAATACTAGACGTCGGCTGTGGTGGAGGAATACTGTCCGAGGGTTTAGCAAAAGCCGGAGCACTCGTGACCGGAATTGACGCAAGTGCAGCTTTAATTATGATAGCAAAGGAACATCGTAATCTCAGTTCAGGTCTACCAAACAGCAACCCTATTTATCACCATAATACTATTGAAGAGCACGTAAAAACACACCCAAACTATTACGATGCAGTCGTTGCTTCAGAAGTTATAGAACACGTGAACAATCCAAAATTATTCCTAAAATCCTGTGTTGAGACTCTCAAATCCGGAGGGGTTATATTTTTAACAACACCAAATAGAACGATATGGTCTTACCTAACTGTTATATTCTTTTGTGAGAAGGTTATATCGATAATACCGAAAGGCGCACACGAATACAGCAAACTGATCACTCCAAGAGAACTTTCCGAAATGCTGACTGAAAATAATTGCAGCGTTGTTTCAAATCAAGGAATTATTCTTAACATAATTACTCGGCACTGGCAATTTATAAAACCACGTAGCTTAATGTATGCAATTCAAGCAGTAAAACTGAATTAA
Protein
MRKIFFHLILVTVIVFWTLIKVPKLLHQYLSERRVTHERKVTECDENSTVIEQNVEIFANLDDWWNPRGKLSTLHKFNVIRIPFIQNGILAHTGINTLNNIKILDVGCGGGILSEGLAKAGALVTGIDASAALIMIAKEHRNLSSGLPNSNPIYHHNTIEEHVKTHPNYYDAVVASEVIEHVNNPKLFLKSCVETLKSGGVIFLTTPNRTIWSYLTVIFFCEKVISIIPKGAHEYSKLITPRELSEMLTENNCSVVSNQGIILNIITRHWQFIKPRSLMYAIQAVKLN
Summary
Description
O-methyltransferase that catalyzes the 2 O-methylation steps in the ubiquinone biosynthetic pathway.
Catalytic Activity
3,4-dihydroxy-5-all-trans-polyprenylbenzoate + S-adenosyl-L-methionine = 3-methoxy,4-hydroxy-5-all-trans-polyprenylbenzoate + H(+) + S-adenosyl-L-homocysteine
a 3-demethylubiquinol + S-adenosyl-L-methionine = a ubiquinol + H(+) + S-adenosyl-L-homocysteine
a 3-demethylubiquinol + S-adenosyl-L-methionine = a ubiquinol + H(+) + S-adenosyl-L-homocysteine
Subunit
Component of a multi-subunit COQ enzyme complex.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. UbiG/COQ3 family.
Uniprot
A0A2H1VUK1
A0A2H1VL37
A0A2W1BI06
A0A2A4JEQ5
A0A212F8D8
H9JPL1
+ More
A0A194RRD2 S4PXP9 A0A1B6D5P4 A0A1B6DMM7 A0A1B6K9N3 A0A1B6K8J6 A0A2H8TEM3 A0A1B6FU63 A0A195E5S0 A0A1B6FXD2 A0A026VXQ4 C4WXE5 A0A0A1XPA7 A0A087ZYC5 A0A2S2P0D0 K7ISW5 A0A151IP29 A0A2S2Q7A1 A0A0V1CKF5 A0A336KST0 A0A067RD09 A0A195FVN1 A0A182HFU4 A0A182LPZ8 Q7Q8W0 A0A182VMK4 A0A182WUT5 A0A0V0S756 A0A195B9G5 A0A026VYR1 A0A151J1B6 A0A182TTG3 A0A0M8ZY76 A0A1B6D2T0 A0A158NJT7 A0A1Y3EH06 A0A195ED63 A0A1A9V9S6 A0A0V1HYG9 A0A084WAV4 A0A0V0XTN1 A0A182QPY8 A0A1B6FCI2 A0A0K8TV91 A0A154PHV5 A0A0K8TTB7 A0A3L8D7D5 A0A151XC01 W5JEG1 A0A158NVH2 A0A1J1J3H8 A0A182MYA7 A0A034WEJ4 A0A2L2YNC2 A0A182MMR4 A0A1B3PDK8 A0A1J1J1P9 A0A182WN26 A0A2L2YN73 A0A1B0CQ57 A0A093C128 A0A131Y3K3 A0A091WR90 A0A1B3PD89 A0A026WC62 A0A0K8R8Y5 A0A0K8WID7 A0A2A3E688 A0A195B7P5 A0A0J7KVZ7 A0A182J4U1 A0A1Y1N1F3 E9HTS6 A0A2P6K0G0 A0A131XRJ0 A0A151WP14 A0A182H3X8 A0A182K681 G1NLE3 A0A0C9QQG2 F4X585 E0VD98 A0A2R5LB16 F6S6G8 A0A023F0Y7 B4N7C3 A0A0A1XS60 A0A1B3PDK7 A0A224YX00 A0A131Z5Y2 A0A0V1EAF9 A0A195E853 A0A1A9ZDD2 A0A195D742
A0A194RRD2 S4PXP9 A0A1B6D5P4 A0A1B6DMM7 A0A1B6K9N3 A0A1B6K8J6 A0A2H8TEM3 A0A1B6FU63 A0A195E5S0 A0A1B6FXD2 A0A026VXQ4 C4WXE5 A0A0A1XPA7 A0A087ZYC5 A0A2S2P0D0 K7ISW5 A0A151IP29 A0A2S2Q7A1 A0A0V1CKF5 A0A336KST0 A0A067RD09 A0A195FVN1 A0A182HFU4 A0A182LPZ8 Q7Q8W0 A0A182VMK4 A0A182WUT5 A0A0V0S756 A0A195B9G5 A0A026VYR1 A0A151J1B6 A0A182TTG3 A0A0M8ZY76 A0A1B6D2T0 A0A158NJT7 A0A1Y3EH06 A0A195ED63 A0A1A9V9S6 A0A0V1HYG9 A0A084WAV4 A0A0V0XTN1 A0A182QPY8 A0A1B6FCI2 A0A0K8TV91 A0A154PHV5 A0A0K8TTB7 A0A3L8D7D5 A0A151XC01 W5JEG1 A0A158NVH2 A0A1J1J3H8 A0A182MYA7 A0A034WEJ4 A0A2L2YNC2 A0A182MMR4 A0A1B3PDK8 A0A1J1J1P9 A0A182WN26 A0A2L2YN73 A0A1B0CQ57 A0A093C128 A0A131Y3K3 A0A091WR90 A0A1B3PD89 A0A026WC62 A0A0K8R8Y5 A0A0K8WID7 A0A2A3E688 A0A195B7P5 A0A0J7KVZ7 A0A182J4U1 A0A1Y1N1F3 E9HTS6 A0A2P6K0G0 A0A131XRJ0 A0A151WP14 A0A182H3X8 A0A182K681 G1NLE3 A0A0C9QQG2 F4X585 E0VD98 A0A2R5LB16 F6S6G8 A0A023F0Y7 B4N7C3 A0A0A1XS60 A0A1B3PDK7 A0A224YX00 A0A131Z5Y2 A0A0V1EAF9 A0A195E853 A0A1A9ZDD2 A0A195D742
Pubmed
28756777
22118469
19121390
26354079
23622113
24508170
+ More
30249741 25830018 20075255 24845553 20966253 12364791 14747013 17210077 21347285 24438588 26369729 20920257 23761445 25348373 26561354 28004739 21292972 28049606 26483478 20838655 21719571 20566863 20431018 25474469 17994087 28797301 26830274
30249741 25830018 20075255 24845553 20966253 12364791 14747013 17210077 21347285 24438588 26369729 20920257 23761445 25348373 26561354 28004739 21292972 28049606 26483478 20838655 21719571 20566863 20431018 25474469 17994087 28797301 26830274
EMBL
ODYU01004521
SOQ44493.1
ODYU01003166
SOQ41569.1
KZ150150
PZC72847.1
+ More
NWSH01001648 PCG70565.1 AGBW02009763 OWR49978.1 BABH01015309 KQ459833 KPJ19865.1 GAIX01004658 JAA87902.1 GEDC01016291 JAS21007.1 GEDC01010357 JAS26941.1 GEBQ01031821 JAT08156.1 GEBQ01032196 JAT07781.1 GFXV01000724 MBW12529.1 GECZ01016044 JAS53725.1 KQ979608 KYN20237.1 GECZ01014914 JAS54855.1 KK107599 QOIP01000012 EZA48588.1 RLU15930.1 ABLF02034105 AK342595 BAH72565.1 GBXI01001577 JAD12715.1 GGMR01010215 MBY22834.1 AAZX01003334 KQ976892 KYN07223.1 GGMS01004268 MBY73471.1 JYDI01000173 KRY49518.1 UFQS01000960 UFQT01000960 SSX07958.1 SSX28192.1 KK852716 KDR17839.1 KQ981215 KYN44518.1 APCN01005583 AAAB01008933 EAA09931.3 JYDL01000030 KRX22558.1 KQ976542 KYM81178.1 EZA48591.1 RLU16367.1 KQ980555 KYN15567.1 KQ435826 KOX72023.1 GEDC01017308 JAS19990.1 ADTU01018253 LVZM01012740 OUC44351.1 KQ979074 KYN22784.1 JYDP01000016 KRZ15763.1 ATLV01022269 KE525331 KFB47348.1 JYDU01000142 KRX91235.1 AXCN02002043 GECZ01021945 JAS47824.1 GDHF01033915 JAI18399.1 KQ434912 KZC11377.1 GDAI01000200 JAI17403.1 RLU16146.1 KQ982316 KYQ57840.1 ADMH02001479 ETN62431.1 ADTU01002888 CVRI01000066 CRL06356.1 GAKP01005833 JAC53119.1 IAAA01030475 LAA09559.1 AXCM01000355 KU660014 AOG17812.1 CRL06373.1 IAAA01030476 LAA09562.1 AJWK01023127 KL451444 KFV05967.1 GEFM01002856 JAP72940.1 KK736148 KFR17635.1 KU659891 AOG17690.1 KK107275 EZA53650.1 GADI01006869 JAA66939.1 GDHF01001505 JAI50809.1 KZ288369 PBC26792.1 KQ976565 KYM80536.1 LBMM01002559 KMQ94662.1 GEZM01020131 JAV89377.1 GL732783 EFX64862.1 MWRG01051081 PRD19819.1 GEFH01000275 JAP68306.1 KQ982893 KYQ49570.1 JXUM01025151 KQ560713 KXJ81190.1 GBYB01002877 JAG72644.1 GL888693 EGI58413.1 AAZO01001263 DS235072 EEB11354.1 GGLE01002588 MBY06714.1 AAMC01037971 AAMC01037972 AAMC01037973 AAMC01037974 AAMC01037975 GBBI01003557 JAC15155.1 CH964182 EDW80264.2 GBXI01000919 JAD13373.1 KU660019 AOG17817.1 GFPF01007206 MAA18352.1 GEDV01003016 JAP85541.1 JYDR01000067 KRY70783.1 KQ979479 KYN21383.1 KQ976750 KYN08696.1
NWSH01001648 PCG70565.1 AGBW02009763 OWR49978.1 BABH01015309 KQ459833 KPJ19865.1 GAIX01004658 JAA87902.1 GEDC01016291 JAS21007.1 GEDC01010357 JAS26941.1 GEBQ01031821 JAT08156.1 GEBQ01032196 JAT07781.1 GFXV01000724 MBW12529.1 GECZ01016044 JAS53725.1 KQ979608 KYN20237.1 GECZ01014914 JAS54855.1 KK107599 QOIP01000012 EZA48588.1 RLU15930.1 ABLF02034105 AK342595 BAH72565.1 GBXI01001577 JAD12715.1 GGMR01010215 MBY22834.1 AAZX01003334 KQ976892 KYN07223.1 GGMS01004268 MBY73471.1 JYDI01000173 KRY49518.1 UFQS01000960 UFQT01000960 SSX07958.1 SSX28192.1 KK852716 KDR17839.1 KQ981215 KYN44518.1 APCN01005583 AAAB01008933 EAA09931.3 JYDL01000030 KRX22558.1 KQ976542 KYM81178.1 EZA48591.1 RLU16367.1 KQ980555 KYN15567.1 KQ435826 KOX72023.1 GEDC01017308 JAS19990.1 ADTU01018253 LVZM01012740 OUC44351.1 KQ979074 KYN22784.1 JYDP01000016 KRZ15763.1 ATLV01022269 KE525331 KFB47348.1 JYDU01000142 KRX91235.1 AXCN02002043 GECZ01021945 JAS47824.1 GDHF01033915 JAI18399.1 KQ434912 KZC11377.1 GDAI01000200 JAI17403.1 RLU16146.1 KQ982316 KYQ57840.1 ADMH02001479 ETN62431.1 ADTU01002888 CVRI01000066 CRL06356.1 GAKP01005833 JAC53119.1 IAAA01030475 LAA09559.1 AXCM01000355 KU660014 AOG17812.1 CRL06373.1 IAAA01030476 LAA09562.1 AJWK01023127 KL451444 KFV05967.1 GEFM01002856 JAP72940.1 KK736148 KFR17635.1 KU659891 AOG17690.1 KK107275 EZA53650.1 GADI01006869 JAA66939.1 GDHF01001505 JAI50809.1 KZ288369 PBC26792.1 KQ976565 KYM80536.1 LBMM01002559 KMQ94662.1 GEZM01020131 JAV89377.1 GL732783 EFX64862.1 MWRG01051081 PRD19819.1 GEFH01000275 JAP68306.1 KQ982893 KYQ49570.1 JXUM01025151 KQ560713 KXJ81190.1 GBYB01002877 JAG72644.1 GL888693 EGI58413.1 AAZO01001263 DS235072 EEB11354.1 GGLE01002588 MBY06714.1 AAMC01037971 AAMC01037972 AAMC01037973 AAMC01037974 AAMC01037975 GBBI01003557 JAC15155.1 CH964182 EDW80264.2 GBXI01000919 JAD13373.1 KU660019 AOG17817.1 GFPF01007206 MAA18352.1 GEDV01003016 JAP85541.1 JYDR01000067 KRY70783.1 KQ979479 KYN21383.1 KQ976750 KYN08696.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053240
UP000078492
UP000053097
+ More
UP000279307 UP000007819 UP000005203 UP000002358 UP000078542 UP000054653 UP000027135 UP000078541 UP000075840 UP000075882 UP000007062 UP000075903 UP000076407 UP000054630 UP000078540 UP000075902 UP000053105 UP000005205 UP000243006 UP000078200 UP000055024 UP000030765 UP000054815 UP000075886 UP000076502 UP000075809 UP000000673 UP000183832 UP000075884 UP000075883 UP000075920 UP000092461 UP000053605 UP000242457 UP000036403 UP000075880 UP000000305 UP000069940 UP000249989 UP000075881 UP000001645 UP000007755 UP000009046 UP000008143 UP000007798 UP000054632 UP000092445
UP000279307 UP000007819 UP000005203 UP000002358 UP000078542 UP000054653 UP000027135 UP000078541 UP000075840 UP000075882 UP000007062 UP000075903 UP000076407 UP000054630 UP000078540 UP000075902 UP000053105 UP000005205 UP000243006 UP000078200 UP000055024 UP000030765 UP000054815 UP000075886 UP000076502 UP000075809 UP000000673 UP000183832 UP000075884 UP000075883 UP000075920 UP000092461 UP000053605 UP000242457 UP000036403 UP000075880 UP000000305 UP000069940 UP000249989 UP000075881 UP000001645 UP000007755 UP000009046 UP000008143 UP000007798 UP000054632 UP000092445
PRIDE
Pfam
Interpro
ProteinModelPortal
A0A2H1VUK1
A0A2H1VL37
A0A2W1BI06
A0A2A4JEQ5
A0A212F8D8
H9JPL1
+ More
A0A194RRD2 S4PXP9 A0A1B6D5P4 A0A1B6DMM7 A0A1B6K9N3 A0A1B6K8J6 A0A2H8TEM3 A0A1B6FU63 A0A195E5S0 A0A1B6FXD2 A0A026VXQ4 C4WXE5 A0A0A1XPA7 A0A087ZYC5 A0A2S2P0D0 K7ISW5 A0A151IP29 A0A2S2Q7A1 A0A0V1CKF5 A0A336KST0 A0A067RD09 A0A195FVN1 A0A182HFU4 A0A182LPZ8 Q7Q8W0 A0A182VMK4 A0A182WUT5 A0A0V0S756 A0A195B9G5 A0A026VYR1 A0A151J1B6 A0A182TTG3 A0A0M8ZY76 A0A1B6D2T0 A0A158NJT7 A0A1Y3EH06 A0A195ED63 A0A1A9V9S6 A0A0V1HYG9 A0A084WAV4 A0A0V0XTN1 A0A182QPY8 A0A1B6FCI2 A0A0K8TV91 A0A154PHV5 A0A0K8TTB7 A0A3L8D7D5 A0A151XC01 W5JEG1 A0A158NVH2 A0A1J1J3H8 A0A182MYA7 A0A034WEJ4 A0A2L2YNC2 A0A182MMR4 A0A1B3PDK8 A0A1J1J1P9 A0A182WN26 A0A2L2YN73 A0A1B0CQ57 A0A093C128 A0A131Y3K3 A0A091WR90 A0A1B3PD89 A0A026WC62 A0A0K8R8Y5 A0A0K8WID7 A0A2A3E688 A0A195B7P5 A0A0J7KVZ7 A0A182J4U1 A0A1Y1N1F3 E9HTS6 A0A2P6K0G0 A0A131XRJ0 A0A151WP14 A0A182H3X8 A0A182K681 G1NLE3 A0A0C9QQG2 F4X585 E0VD98 A0A2R5LB16 F6S6G8 A0A023F0Y7 B4N7C3 A0A0A1XS60 A0A1B3PDK7 A0A224YX00 A0A131Z5Y2 A0A0V1EAF9 A0A195E853 A0A1A9ZDD2 A0A195D742
A0A194RRD2 S4PXP9 A0A1B6D5P4 A0A1B6DMM7 A0A1B6K9N3 A0A1B6K8J6 A0A2H8TEM3 A0A1B6FU63 A0A195E5S0 A0A1B6FXD2 A0A026VXQ4 C4WXE5 A0A0A1XPA7 A0A087ZYC5 A0A2S2P0D0 K7ISW5 A0A151IP29 A0A2S2Q7A1 A0A0V1CKF5 A0A336KST0 A0A067RD09 A0A195FVN1 A0A182HFU4 A0A182LPZ8 Q7Q8W0 A0A182VMK4 A0A182WUT5 A0A0V0S756 A0A195B9G5 A0A026VYR1 A0A151J1B6 A0A182TTG3 A0A0M8ZY76 A0A1B6D2T0 A0A158NJT7 A0A1Y3EH06 A0A195ED63 A0A1A9V9S6 A0A0V1HYG9 A0A084WAV4 A0A0V0XTN1 A0A182QPY8 A0A1B6FCI2 A0A0K8TV91 A0A154PHV5 A0A0K8TTB7 A0A3L8D7D5 A0A151XC01 W5JEG1 A0A158NVH2 A0A1J1J3H8 A0A182MYA7 A0A034WEJ4 A0A2L2YNC2 A0A182MMR4 A0A1B3PDK8 A0A1J1J1P9 A0A182WN26 A0A2L2YN73 A0A1B0CQ57 A0A093C128 A0A131Y3K3 A0A091WR90 A0A1B3PD89 A0A026WC62 A0A0K8R8Y5 A0A0K8WID7 A0A2A3E688 A0A195B7P5 A0A0J7KVZ7 A0A182J4U1 A0A1Y1N1F3 E9HTS6 A0A2P6K0G0 A0A131XRJ0 A0A151WP14 A0A182H3X8 A0A182K681 G1NLE3 A0A0C9QQG2 F4X585 E0VD98 A0A2R5LB16 F6S6G8 A0A023F0Y7 B4N7C3 A0A0A1XS60 A0A1B3PDK7 A0A224YX00 A0A131Z5Y2 A0A0V1EAF9 A0A195E853 A0A1A9ZDD2 A0A195D742
PDB
4KDC
E-value=6.60437e-31,
Score=333
Ontologies
PATHWAY
GO
PANTHER
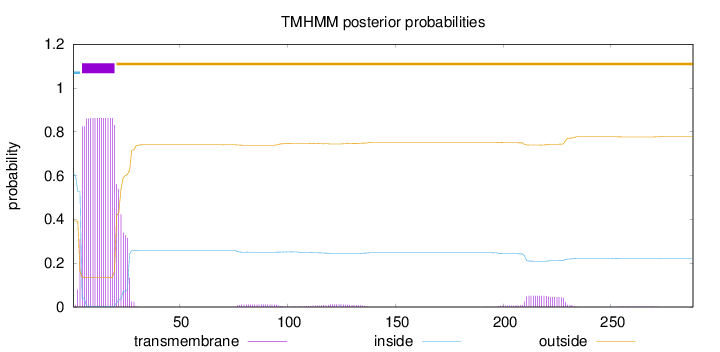
Topology
Subcellular location
Mitochondrion inner membrane
Length:
288
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.32617
Exp number, first 60 AAs:
16.78392
Total prob of N-in:
0.60701
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 20
outside
21 - 288
Population Genetic Test Statistics
Pi
206.042587
Theta
179.111478
Tajima's D
0.280443
CLR
0.697618
CSRT
0.449077546122694
Interpretation
Uncertain
