Gene
KWMTBOMO13801
Pre Gene Modal
BGIBMGA011544
Annotation
PREDICTED:_uncharacterized_aarF_domain-containing_protein_kinase_1_[Papilio_xuthus]
Full name
Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial
Location in the cell
Cytoplasmic Reliability : 3.07
Sequence
CDS
ATGTTCTCCAGAAAGTTGACTAGGATCATAAAATACGGCTTTTATGGTGGTGTTGCTGTTGGTGGTTGTGTAACTGCTTTAAAATTTCATGATGGTGATTATAATTCGTTAGCCATCGTCAGAATCACGAGGACAGCATGTACTGCTGTGGACATAGGCAGGACTTACCAGTCTTTGTTGTACAGCAAAGATTGGGATAGATCGTCCCCCGAGTATTTAGAAGTGAAGAAACAAGCTCATCAAATTGGGGCAGAGAAACTGCTTGAATTATGCAAAGCTAACAAAGGTGTGTACATTAAAGTTGGACAGCACGTAGGCGCGTTAGATTACTTGTTGCCTACTGAATATGTCAAAACAATGCGTGTTTTGCATAAAGATGCGCCTCAAAACACAGTCGAAGAACTATACAATGTCATCAGGGAAGATTTGAAAAAGGATCCAAAGGAACTGTTTGACGACTTTGAACCTGAGCCTCTGGGCACAGCGTCACTAGCACAAGTTCATAAAGCAAAATTAAAAGATGGTACTGAAGTTGCGGTCAAAGTACAACATCATTTTGTGAGGAAAAATATTAAGATTGACTTAAAATGGATGGAATTTATTATAAACACAATGTCAAAAGTATTTCCTGACTTCCAAATGCAATGGCTAGTGGATGAAACGAAGAAAAACATTGCAAAAGAGCTGGATTTCCTGCAAGAGGGTCAAAATGCAGAGAAGGTAGCTGAATTGTTCAGTAATTACAAATGGCTGAAGGTTCCAAAGATTTTCTGGGATTACAGTACAGAGAGAGTGCTGGTCATGGAATATGTTTACGGTGGACAAGTTAATGATGTTAATTATATTGATACACATAAAATCAACAGATATGATTTATGTAAGAAGCTTGGTGATCTCTACTCCCACATGATCTTCATAACTGGTTTTGTGCACAGTGATCCACATCCAGGGAATATACTGGTTAAAAAAGCTCCAGGAGACAAGGATGTAACAGTTTATTTGCTGGACCATGGGTTATATGCTCAACTTTCGGATAAATTCAGATATCACTATTCAAAATTGTGGCTATCCATTATAGACAGAAATCGTCAAGAAATGAAAGTACATGCAGAGCAGCTAGGTATTCGAAATGAATTGTATGGTTTGTTTGCTTGTATGGTAACGGGTAGACCTTGGGATTCTATTATGAAAGGAATTGGCAAAACTAAGCCATCTTCAGAAGAGAAATCAACTTTTCAAAACGAATTGCCTAACATCCTCCACCATATCACTGATTGCCTGGAGCATGTGGACCGGCAAGCATTATTGGTTCTCAAAACCAATGATTTAATCCGTAGCATAGAATATGCTCTAGGAACACAAGAGAGAATGTGTGGTTTTGTTGTAATGAGCAAATGCTGCATACAAAGCGCATATAGAGAAGAAATTAACAAAGAGAAGTCTATATTTAGAAGACAATACTTAAATTTAAAATTTAGTTGGACCATGCTGGTGCTTTACTTGTACAGTGTATATTTAAATTTATGTGATACATTAATTTTATAA
Protein
MFSRKLTRIIKYGFYGGVAVGGCVTALKFHDGDYNSLAIVRITRTACTAVDIGRTYQSLLYSKDWDRSSPEYLEVKKQAHQIGAEKLLELCKANKGVYIKVGQHVGALDYLLPTEYVKTMRVLHKDAPQNTVEELYNVIREDLKKDPKELFDDFEPEPLGTASLAQVHKAKLKDGTEVAVKVQHHFVRKNIKIDLKWMEFIINTMSKVFPDFQMQWLVDETKKNIAKELDFLQEGQNAEKVAELFSNYKWLKVPKIFWDYSTERVLVMEYVYGGQVNDVNYIDTHKINRYDLCKKLGDLYSHMIFITGFVHSDPHPGNILVKKAPGDKDVTVYLLDHGLYAQLSDKFRYHYSKLWLSIIDRNRQEMKVHAEQLGIRNELYGLFACMVTGRPWDSIMKGIGKTKPSSEEKSTFQNELPNILHHITDCLEHVDRQALLVLKTNDLIRSIEYALGTQERMCGFVVMSKCCIQSAYREEINKEKSIFRRQYLNLKFSWTMLVLYLYSVYLNLCDTLIL
Summary
Description
FAD-dependent monooxygenase required for the C5-ring hydroxylation during ubiquinone biosynthesis. Catalyzes the hydroxylation of 3-polyprenyl-4-hydroxybenzoic acid to 3-polyprenyl-4,5-dihydroxybenzoic acid. The electrons required for the hydroxylation reaction may be funneled indirectly from NADPH via a ferredoxin/ferredoxin reductase system to COQ6.
Cofactor
FAD
Subunit
Component of a multi-subunit COQ enzyme complex.
Similarity
Belongs to the UbiH/COQ6 family.
Uniprot
H9JPT9
A0A2W1BC34
A0A2A4JES8
S4P7Q4
A0A194RPT1
A0A0N1IC13
+ More
A0A3S2LCC9 A0A212EN49 B4KSZ2 B4LML0 A0A1B0GQQ8 Q174U0 A0A0M3QUL2 B4MQI3 A0A1I8PXL0 Q28XA0 B4H573 A0A0A1WTQ8 A0A1I8NBD7 B4QCH3 A0A1W4W0J2 Q9W133 T1PBJ4 B4PBE5 B3NQW0 A0A182HE87 A0A0K8UD15 A0A3B0JNU3 A0A1B0CL95 B0WTN9 B4IH55 B3MEA3 W8BUU5 D7EID2 A0A182Y1T3 A0A1Q3FDJ0 A0A2M4AFN4 A0A182R5E6 A0A182PBR2 A0A182V7L8 A0A084WHW7 A0A1B0AZL7 A0A0L0C2K8 A0A1A9ZE16 A0A182FU32 A0A182XD70 A0A182MYJ6 A0A182KSN2 A0A182UBI8 A0A182HQ26 A0A2M4BKF9 A0A1A9VID0 W5JBQ8 A0A2M4BKD6 Q7Q071 A0A182SHR8 A0A182Q9J0 A0A1B0FJQ8 A0A182JUX9 A0A182JC41 A0A1A9W9R5 A0A1A9YE92 A0A1L8DN33 A0A182W965 A0A182MM32 U4U281 N6UFA2 A0A067RDK5 B4J4X4 E2APB4 A0A1B6D1A7 A0A195C072 A0A154P4Z7 A0A0J7KY00 A0A0C9PPS8 A0A0C9QI43 A0A310SM22 A0A0L7QXF6 A0A088A362 A0A195FEZ9 A0A2A3E8X7 A0A195AWY0 A0A195EH71 A0A158NHX4 A0A1W4WI66 E2C8K7 A0A2J7QSZ4 E9JB45 A0A1Y1MAX0 A0A026WIF8 A0A1B6MKA0 A0A1B6INZ9 A0A0M9A7E4 A0A023F2D9 A0A1W4WUC7 A0A3Q0IXR1 A0A1B0CL93 E0VQ85 F4WPR5
A0A3S2LCC9 A0A212EN49 B4KSZ2 B4LML0 A0A1B0GQQ8 Q174U0 A0A0M3QUL2 B4MQI3 A0A1I8PXL0 Q28XA0 B4H573 A0A0A1WTQ8 A0A1I8NBD7 B4QCH3 A0A1W4W0J2 Q9W133 T1PBJ4 B4PBE5 B3NQW0 A0A182HE87 A0A0K8UD15 A0A3B0JNU3 A0A1B0CL95 B0WTN9 B4IH55 B3MEA3 W8BUU5 D7EID2 A0A182Y1T3 A0A1Q3FDJ0 A0A2M4AFN4 A0A182R5E6 A0A182PBR2 A0A182V7L8 A0A084WHW7 A0A1B0AZL7 A0A0L0C2K8 A0A1A9ZE16 A0A182FU32 A0A182XD70 A0A182MYJ6 A0A182KSN2 A0A182UBI8 A0A182HQ26 A0A2M4BKF9 A0A1A9VID0 W5JBQ8 A0A2M4BKD6 Q7Q071 A0A182SHR8 A0A182Q9J0 A0A1B0FJQ8 A0A182JUX9 A0A182JC41 A0A1A9W9R5 A0A1A9YE92 A0A1L8DN33 A0A182W965 A0A182MM32 U4U281 N6UFA2 A0A067RDK5 B4J4X4 E2APB4 A0A1B6D1A7 A0A195C072 A0A154P4Z7 A0A0J7KY00 A0A0C9PPS8 A0A0C9QI43 A0A310SM22 A0A0L7QXF6 A0A088A362 A0A195FEZ9 A0A2A3E8X7 A0A195AWY0 A0A195EH71 A0A158NHX4 A0A1W4WI66 E2C8K7 A0A2J7QSZ4 E9JB45 A0A1Y1MAX0 A0A026WIF8 A0A1B6MKA0 A0A1B6INZ9 A0A0M9A7E4 A0A023F2D9 A0A1W4WUC7 A0A3Q0IXR1 A0A1B0CL93 E0VQ85 F4WPR5
EC Number
1.14.13.-
Pubmed
19121390
28756777
23622113
26354079
22118469
17994087
+ More
18057021 17510324 15632085 25830018 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 24495485 18362917 19820115 25244985 24438588 26108605 20966253 20920257 23761445 12364791 14747013 17210077 23537049 24845553 20798317 21347285 21282665 28004739 24508170 30249741 25474469 20566863 21719571
18057021 17510324 15632085 25830018 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 24495485 18362917 19820115 25244985 24438588 26108605 20966253 20920257 23761445 12364791 14747013 17210077 23537049 24845553 20798317 21347285 21282665 28004739 24508170 30249741 25474469 20566863 21719571
EMBL
BABH01015307
BABH01015308
BABH01015309
KZ150150
PZC72849.1
NWSH01001648
+ More
PCG70567.1 GAIX01004489 JAA88071.1 KQ459833 KPJ19863.1 KQ459037 KPJ04297.1 RSAL01000242 RVE43640.1 AGBW02013730 OWR42906.1 CH933808 EDW08489.1 CH940648 EDW59997.1 KRF79188.1 KRF79189.1 AJVK01007787 CH477406 EAT41561.1 CP012524 ALC40823.1 CH963849 EDW74372.1 CM000071 EAL26416.1 CH479210 EDW32909.1 GBXI01011838 JAD02454.1 CM000362 CM002911 EDX08589.1 KMY96393.1 AE013599 AY118660 AAF47244.2 AAM50029.1 AHN56646.1 KA646039 AFP60668.1 CM000158 EDW92577.1 CH954179 EDV57043.1 JXUM01035328 KQ561055 KXJ79848.1 GDHF01027742 JAI24572.1 OUUW01000001 SPP75006.1 AJWK01017050 DS232090 EDS34497.1 CH480838 EDW49231.1 CH902619 EDV36509.1 KPU76265.1 KPU76266.1 GAMC01005821 GAMC01005820 GAMC01005819 GAMC01005818 GAMC01005817 JAC00739.1 DS497676 EFA11786.1 GFDL01009422 JAV25623.1 GGFK01006252 MBW39573.1 ATLV01023892 KE525347 KFB49811.1 JXJN01006363 JRES01000975 KNC26560.1 APCN01003288 GGFJ01004396 MBW53537.1 ADMH02001693 ETN61401.1 GGFJ01004395 MBW53536.1 AAAB01008986 EAA00298.2 AXCN02001041 CCAG010016144 AXCP01006766 GFDF01006253 JAV07831.1 AXCM01014079 KB631815 ERL86393.1 APGK01029188 KB740727 ENN79306.1 KK852771 KDR16899.1 CH916367 EDW01680.1 GL441504 EFN64713.1 GEDC01017841 JAS19457.1 KQ978501 KYM93566.1 KQ434809 KZC06394.1 LBMM01002197 KMQ95158.1 GBYB01003208 JAG72975.1 GBYB01003209 JAG72976.1 KQ761039 OAD58345.1 KQ414704 KOC63290.1 KQ981636 KYN38797.1 KZ288322 PBC28177.1 KQ976725 KYM76555.1 KQ978957 KYN27222.1 ADTU01016101 GL453666 EFN75737.1 NEVH01011202 PNF31716.1 GL770938 EFZ09920.1 GEZM01036375 JAV82751.1 KK107185 QOIP01000012 EZA55832.1 RLU16344.1 GEBQ01003597 JAT36380.1 GECU01019048 GECU01017567 JAS88658.1 JAS90139.1 KQ435719 KOX78760.1 GBBI01003267 JAC15445.1 AJWK01017049 DS235402 EEB15541.1 GL888255 EGI63806.1
PCG70567.1 GAIX01004489 JAA88071.1 KQ459833 KPJ19863.1 KQ459037 KPJ04297.1 RSAL01000242 RVE43640.1 AGBW02013730 OWR42906.1 CH933808 EDW08489.1 CH940648 EDW59997.1 KRF79188.1 KRF79189.1 AJVK01007787 CH477406 EAT41561.1 CP012524 ALC40823.1 CH963849 EDW74372.1 CM000071 EAL26416.1 CH479210 EDW32909.1 GBXI01011838 JAD02454.1 CM000362 CM002911 EDX08589.1 KMY96393.1 AE013599 AY118660 AAF47244.2 AAM50029.1 AHN56646.1 KA646039 AFP60668.1 CM000158 EDW92577.1 CH954179 EDV57043.1 JXUM01035328 KQ561055 KXJ79848.1 GDHF01027742 JAI24572.1 OUUW01000001 SPP75006.1 AJWK01017050 DS232090 EDS34497.1 CH480838 EDW49231.1 CH902619 EDV36509.1 KPU76265.1 KPU76266.1 GAMC01005821 GAMC01005820 GAMC01005819 GAMC01005818 GAMC01005817 JAC00739.1 DS497676 EFA11786.1 GFDL01009422 JAV25623.1 GGFK01006252 MBW39573.1 ATLV01023892 KE525347 KFB49811.1 JXJN01006363 JRES01000975 KNC26560.1 APCN01003288 GGFJ01004396 MBW53537.1 ADMH02001693 ETN61401.1 GGFJ01004395 MBW53536.1 AAAB01008986 EAA00298.2 AXCN02001041 CCAG010016144 AXCP01006766 GFDF01006253 JAV07831.1 AXCM01014079 KB631815 ERL86393.1 APGK01029188 KB740727 ENN79306.1 KK852771 KDR16899.1 CH916367 EDW01680.1 GL441504 EFN64713.1 GEDC01017841 JAS19457.1 KQ978501 KYM93566.1 KQ434809 KZC06394.1 LBMM01002197 KMQ95158.1 GBYB01003208 JAG72975.1 GBYB01003209 JAG72976.1 KQ761039 OAD58345.1 KQ414704 KOC63290.1 KQ981636 KYN38797.1 KZ288322 PBC28177.1 KQ976725 KYM76555.1 KQ978957 KYN27222.1 ADTU01016101 GL453666 EFN75737.1 NEVH01011202 PNF31716.1 GL770938 EFZ09920.1 GEZM01036375 JAV82751.1 KK107185 QOIP01000012 EZA55832.1 RLU16344.1 GEBQ01003597 JAT36380.1 GECU01019048 GECU01017567 JAS88658.1 JAS90139.1 KQ435719 KOX78760.1 GBBI01003267 JAC15445.1 AJWK01017049 DS235402 EEB15541.1 GL888255 EGI63806.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000009192 UP000008792 UP000092462 UP000008820 UP000092553 UP000007798 UP000095300 UP000001819 UP000008744 UP000095301 UP000000304 UP000192221 UP000000803 UP000002282 UP000008711 UP000069940 UP000249989 UP000268350 UP000092461 UP000002320 UP000001292 UP000007801 UP000007266 UP000076408 UP000075900 UP000075885 UP000075903 UP000030765 UP000092460 UP000037069 UP000092445 UP000069272 UP000076407 UP000075884 UP000075882 UP000075902 UP000075840 UP000078200 UP000000673 UP000007062 UP000075901 UP000075886 UP000092444 UP000075881 UP000075880 UP000091820 UP000092443 UP000075920 UP000075883 UP000030742 UP000019118 UP000027135 UP000001070 UP000000311 UP000078542 UP000076502 UP000036403 UP000053825 UP000005203 UP000078541 UP000242457 UP000078540 UP000078492 UP000005205 UP000192223 UP000008237 UP000235965 UP000053097 UP000279307 UP000053105 UP000079169 UP000009046 UP000007755
UP000009192 UP000008792 UP000092462 UP000008820 UP000092553 UP000007798 UP000095300 UP000001819 UP000008744 UP000095301 UP000000304 UP000192221 UP000000803 UP000002282 UP000008711 UP000069940 UP000249989 UP000268350 UP000092461 UP000002320 UP000001292 UP000007801 UP000007266 UP000076408 UP000075900 UP000075885 UP000075903 UP000030765 UP000092460 UP000037069 UP000092445 UP000069272 UP000076407 UP000075884 UP000075882 UP000075902 UP000075840 UP000078200 UP000000673 UP000007062 UP000075901 UP000075886 UP000092444 UP000075881 UP000075880 UP000091820 UP000092443 UP000075920 UP000075883 UP000030742 UP000019118 UP000027135 UP000001070 UP000000311 UP000078542 UP000076502 UP000036403 UP000053825 UP000005203 UP000078541 UP000242457 UP000078540 UP000078492 UP000005205 UP000192223 UP000008237 UP000235965 UP000053097 UP000279307 UP000053105 UP000079169 UP000009046 UP000007755
Interpro
IPR004147
UbiB_dom
+ More
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR018488 cNMP-bd_CS
IPR012541 DBP10_C
IPR027417 P-loop_NTPase
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR036188 FAD/NAD-bd_sf
IPR000689 UbQ_mOase_COQ6
IPR002938 FAD-bd
IPR018168 Ubi_Hdrlase_CS
IPR010971 UbiH/COQ6
IPR006311 TAT_signal
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR018488 cNMP-bd_CS
IPR012541 DBP10_C
IPR027417 P-loop_NTPase
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR036188 FAD/NAD-bd_sf
IPR000689 UbQ_mOase_COQ6
IPR002938 FAD-bd
IPR018168 Ubi_Hdrlase_CS
IPR010971 UbiH/COQ6
IPR006311 TAT_signal
Gene 3D
CDD
ProteinModelPortal
H9JPT9
A0A2W1BC34
A0A2A4JES8
S4P7Q4
A0A194RPT1
A0A0N1IC13
+ More
A0A3S2LCC9 A0A212EN49 B4KSZ2 B4LML0 A0A1B0GQQ8 Q174U0 A0A0M3QUL2 B4MQI3 A0A1I8PXL0 Q28XA0 B4H573 A0A0A1WTQ8 A0A1I8NBD7 B4QCH3 A0A1W4W0J2 Q9W133 T1PBJ4 B4PBE5 B3NQW0 A0A182HE87 A0A0K8UD15 A0A3B0JNU3 A0A1B0CL95 B0WTN9 B4IH55 B3MEA3 W8BUU5 D7EID2 A0A182Y1T3 A0A1Q3FDJ0 A0A2M4AFN4 A0A182R5E6 A0A182PBR2 A0A182V7L8 A0A084WHW7 A0A1B0AZL7 A0A0L0C2K8 A0A1A9ZE16 A0A182FU32 A0A182XD70 A0A182MYJ6 A0A182KSN2 A0A182UBI8 A0A182HQ26 A0A2M4BKF9 A0A1A9VID0 W5JBQ8 A0A2M4BKD6 Q7Q071 A0A182SHR8 A0A182Q9J0 A0A1B0FJQ8 A0A182JUX9 A0A182JC41 A0A1A9W9R5 A0A1A9YE92 A0A1L8DN33 A0A182W965 A0A182MM32 U4U281 N6UFA2 A0A067RDK5 B4J4X4 E2APB4 A0A1B6D1A7 A0A195C072 A0A154P4Z7 A0A0J7KY00 A0A0C9PPS8 A0A0C9QI43 A0A310SM22 A0A0L7QXF6 A0A088A362 A0A195FEZ9 A0A2A3E8X7 A0A195AWY0 A0A195EH71 A0A158NHX4 A0A1W4WI66 E2C8K7 A0A2J7QSZ4 E9JB45 A0A1Y1MAX0 A0A026WIF8 A0A1B6MKA0 A0A1B6INZ9 A0A0M9A7E4 A0A023F2D9 A0A1W4WUC7 A0A3Q0IXR1 A0A1B0CL93 E0VQ85 F4WPR5
A0A3S2LCC9 A0A212EN49 B4KSZ2 B4LML0 A0A1B0GQQ8 Q174U0 A0A0M3QUL2 B4MQI3 A0A1I8PXL0 Q28XA0 B4H573 A0A0A1WTQ8 A0A1I8NBD7 B4QCH3 A0A1W4W0J2 Q9W133 T1PBJ4 B4PBE5 B3NQW0 A0A182HE87 A0A0K8UD15 A0A3B0JNU3 A0A1B0CL95 B0WTN9 B4IH55 B3MEA3 W8BUU5 D7EID2 A0A182Y1T3 A0A1Q3FDJ0 A0A2M4AFN4 A0A182R5E6 A0A182PBR2 A0A182V7L8 A0A084WHW7 A0A1B0AZL7 A0A0L0C2K8 A0A1A9ZE16 A0A182FU32 A0A182XD70 A0A182MYJ6 A0A182KSN2 A0A182UBI8 A0A182HQ26 A0A2M4BKF9 A0A1A9VID0 W5JBQ8 A0A2M4BKD6 Q7Q071 A0A182SHR8 A0A182Q9J0 A0A1B0FJQ8 A0A182JUX9 A0A182JC41 A0A1A9W9R5 A0A1A9YE92 A0A1L8DN33 A0A182W965 A0A182MM32 U4U281 N6UFA2 A0A067RDK5 B4J4X4 E2APB4 A0A1B6D1A7 A0A195C072 A0A154P4Z7 A0A0J7KY00 A0A0C9PPS8 A0A0C9QI43 A0A310SM22 A0A0L7QXF6 A0A088A362 A0A195FEZ9 A0A2A3E8X7 A0A195AWY0 A0A195EH71 A0A158NHX4 A0A1W4WI66 E2C8K7 A0A2J7QSZ4 E9JB45 A0A1Y1MAX0 A0A026WIF8 A0A1B6MKA0 A0A1B6INZ9 A0A0M9A7E4 A0A023F2D9 A0A1W4WUC7 A0A3Q0IXR1 A0A1B0CL93 E0VQ85 F4WPR5
PDB
5YK2
E-value=9.8125e-25,
Score=282
Ontologies
GO
Topology
Subcellular location
Mitochondrion inner membrane
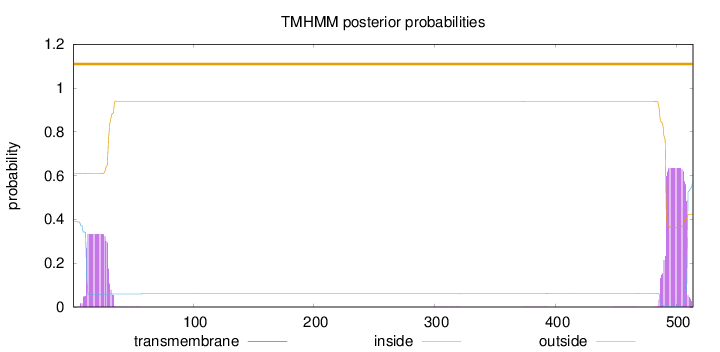
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
18.68416
Exp number, first 60 AAs:
6.51053
Total prob of N-in:
0.38918
outside
1 - 514
Population Genetic Test Statistics
Pi
272.658804
Theta
204.377697
Tajima's D
1.30258
CLR
0
CSRT
0.741062946852657
Interpretation
Uncertain
