Gene
KWMTBOMO13799 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011543
Annotation
dicer-2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.708 Golgi Reliability : 1.633
Sequence
CDS
ATGGGGATTCGTCCAACATTCTTAATCGGCATACAGTTCACCGAAGAGGAAGTAATGACCGGTGAATTAAGTGAAGAAAGCTGGGAATCTGTGATGAATAATTTCCAAAATGGCATCGCTGAGGCAGAACCCGAAGGCTGTGCCCAAAATTCTATGCAATGCTATGTACATTCACAAGCAGTTGCGGACAAATCTATAGCCGATTCCGTGGAAGCTCTGATCGGCACATATCTACTTAGTGGAGGGATATTAGCAGCTGTAAAACTACTTGAATGGATGGAGGTGTTTCCGCCACAGGATAACTTTGCAGACATGCTCCACAAGCCGGTGCAAACACCGTTATCGAAGAATTTAGCTACCGAAGCTGATATTGACTTTTTACTAAACAATTCGAGAACTGATGTCGAAAAGATCCTAAACTACACGTTCAAAGATTCAACCTTCCTGCTAAACGCTCTGTCTCACTCGTCGTACATCCGGAACAGGCTGACGAGCTCGTACGAGCGCCTCGAGTTCCTCGGGGACGCCATAATCGATTTCCTGGTGACGTCACACATCTTCGAGAACTGTCGCGAACTGAAGCCCGGCGAAATGACTGACCTGAGATCCGCCCTCGTCAATAACGTCACGTTCGCGGCCTACACCGTCAAACTTGGATTGCACAAGTACCTCTGCTCGGAGCTGAACCCTTCGTTGGAGAAGGCGATAATGAAATTCGTCGAACATCAAATAGAAAGAAATCACGAGATTGAGTTCGACGTTCTGTTACTTATCAGCGAAGTCGATTGTCAAGTCGCTGAATATGTTGATGTGCCGAAGGTGTTGAGCGACATTTTCGAAGCATTAGTCGGCGCCGTGTACTTGGACTCGGGCGGGAATCTGGAGACGGTGTGGTCGGTTCTGTACAAGATAATGCACGTCGAGATCGATTCGTTCTCGAAACGCATACCGAAGCAGCCCGTCAAAATTCTATACGAAATGATACACGCTTGTCCGGTTTTCGAAAAACAAATAGTCGTCAATCCGGACATCCCGAAGGTAATGGTGCCGGTTTCGTTCACGAAGAACGACCAGCGTCTCGCCGCCTACGGAGTCGGCGTCAACAAATCGCAAGCGAAGAGGGCCGCCGCGAAATTAGCTCTCAAAATATTAGACTCTTAA
Protein
MGIRPTFLIGIQFTEEEVMTGELSEESWESVMNNFQNGIAEAEPEGCAQNSMQCYVHSQAVADKSIADSVEALIGTYLLSGGILAAVKLLEWMEVFPPQDNFADMLHKPVQTPLSKNLATEADIDFLLNNSRTDVEKILNYTFKDSTFLLNALSHSSYIRNRLTSSYERLEFLGDAIIDFLVTSHIFENCRELKPGEMTDLRSALVNNVTFAAYTVKLGLHKYLCSELNPSLEKAIMKFVEHQIERNHEIEFDVLLLISEVDCQVAEYVDVPKVLSDIFEALVGAVYLDSGGNLETVWSVLYKIMHVEIDSFSKRIPKQPVKILYEMIHACPVFEKQIVVNPDIPKVMVPVSFTKNDQRLAAYGVGVNKSQAKRAAAKLALKILDS
Summary
Similarity
Belongs to the helicase family. Dicer subfamily.
Uniprot
H9JPT8
D7UT11
A0A2A4JFH8
A0A3S2LSR5
V9VKP3
A0A0N1IDN4
+ More
A0A2P1DM58 A0A2W1BBL1 A0A212EN34 A0A2H1WFS6 A0A194RUP4 A0A1W4X5J6 A0A0B5G8Z8 A0A0T6B8D4 A0A2P1DM78 K0JC91 A0A2A3E7Y1 A0A0H4SZH3 D6WA66 A0A1Y1MHP6 A0A386QX51 A0A0H4T2N5 A0A0M8ZTS6 A0A088ABX2 A0A1Y1MD92 A0A140H4W2 A0A1Y1MIC1 K9LIV6 E0VG65 A0A1Y1MF64 A0A1B6I6C5 A0A1B6I9V0 A0A2J7PPT2 E2AVZ8 A0A151X4G5 A0A026WZU4 A0A1B6H3B9 A0A3L8DUT7 A0A0S1RSS9 A0A1Y1MCU9 L7Z6F8 A0A1W7HGU9 A0A1W7HGU4 N6UUA9 N6TTD5 A0A154PHY9 U4UQA5 A0A195BR89 A0A158NDW6 A0A0C9QFQ7 A0A0C9RZ40 F4W7V0 A0A1B6FF57 A0A069DXQ4 A0A195D7X2 K7YZ50 A0A2P1DM48 A0A2I6PIZ5 K7J5H5 A0A195ESA7 A0A232F391 B0WTP2 A0A224X595 A0A1L3A5U6 A0A1B6DNE2 A0A023F657 A0A1Q3FBT5 A0A1Q3FBL4 A0A0J7L2K8 A0A1L7NZN0 A0A060H2K4 A0A182GUS7 A0A0A7DVT8 E2BY57 A0A2M4AB33 A0A2M4ABM6 A0A1L8DXZ8 Q174T8 H2BNN0 Q49LL4 W4VRF8 A0A182FU44 A0A182HE85 A0A1W7HGU5 H2BNN1 H2BNM7 W5JN74 A0A0A9YBL6 A0A146M220 A0A0K8S5D2 A0A0A9Y1K7 A0A182PBR4 A0A2R7VQG7 A0A226DKF2 A0A182PZN5 A0A182Y1T1 A0A2S2PGD2 A0A2S2NCI3
A0A2P1DM58 A0A2W1BBL1 A0A212EN34 A0A2H1WFS6 A0A194RUP4 A0A1W4X5J6 A0A0B5G8Z8 A0A0T6B8D4 A0A2P1DM78 K0JC91 A0A2A3E7Y1 A0A0H4SZH3 D6WA66 A0A1Y1MHP6 A0A386QX51 A0A0H4T2N5 A0A0M8ZTS6 A0A088ABX2 A0A1Y1MD92 A0A140H4W2 A0A1Y1MIC1 K9LIV6 E0VG65 A0A1Y1MF64 A0A1B6I6C5 A0A1B6I9V0 A0A2J7PPT2 E2AVZ8 A0A151X4G5 A0A026WZU4 A0A1B6H3B9 A0A3L8DUT7 A0A0S1RSS9 A0A1Y1MCU9 L7Z6F8 A0A1W7HGU9 A0A1W7HGU4 N6UUA9 N6TTD5 A0A154PHY9 U4UQA5 A0A195BR89 A0A158NDW6 A0A0C9QFQ7 A0A0C9RZ40 F4W7V0 A0A1B6FF57 A0A069DXQ4 A0A195D7X2 K7YZ50 A0A2P1DM48 A0A2I6PIZ5 K7J5H5 A0A195ESA7 A0A232F391 B0WTP2 A0A224X595 A0A1L3A5U6 A0A1B6DNE2 A0A023F657 A0A1Q3FBT5 A0A1Q3FBL4 A0A0J7L2K8 A0A1L7NZN0 A0A060H2K4 A0A182GUS7 A0A0A7DVT8 E2BY57 A0A2M4AB33 A0A2M4ABM6 A0A1L8DXZ8 Q174T8 H2BNN0 Q49LL4 W4VRF8 A0A182FU44 A0A182HE85 A0A1W7HGU5 H2BNN1 H2BNM7 W5JN74 A0A0A9YBL6 A0A146M220 A0A0K8S5D2 A0A0A9Y1K7 A0A182PBR4 A0A2R7VQG7 A0A226DKF2 A0A182PZN5 A0A182Y1T1 A0A2S2PGD2 A0A2S2NCI3
Pubmed
19121390
26354079
28756777
22118469
25541004
22562088
+ More
18362917 19820115 28004739 30261185 26864546 20566863 20798317 24508170 30249741 23029445 23537049 21347285 21719571 26334808 23232437 29267401 20075255 28648823 25474469 28034629 24391810 26483478 17510324 20920257 23761445 25401762 26823975 25244985
18362917 19820115 28004739 30261185 26864546 20566863 20798317 24508170 30249741 23029445 23537049 21347285 21719571 26334808 23232437 29267401 20075255 28648823 25474469 28034629 24391810 26483478 17510324 20920257 23761445 25401762 26823975 25244985
EMBL
BABH01015301
AB566386
BAJ11655.1
NWSH01001648
PCG70569.1
RSAL01000242
+ More
RVE43638.1 KF717092 AHC98017.1 KQ459037 KPJ04295.1 MG225431 AVK59442.1 KZ150192 PZC72388.1 AGBW02013730 OWR42904.1 ODYU01008375 SOQ51911.1 KQ459833 KPJ19861.1 KP036490 AJF15703.1 LJIG01009215 KRT83525.1 MG225447 AVK59458.1 HE647851 CCF23094.1 KZ288338 PBC27815.1 KP230552 AKQ00041.1 KQ971312 EEZ99277.1 GEZM01034249 JAV83855.1 MH170281 AYE54589.1 KP230553 AKQ00042.1 KQ435851 KOX70787.1 GEZM01034726 GEZM01034725 JAV83643.1 KT191018 AMN92163.1 GEZM01034724 JAV83646.1 JQ037907 AFK73581.1 DS235131 EEB12371.1 GEZM01034723 JAV83648.1 GECU01025228 JAS82478.1 GECU01024018 JAS83688.1 NEVH01022647 PNF18316.1 GL443213 EFN62420.1 KQ982548 KYQ55277.1 KK107054 EZA61552.1 GECZ01000596 JAS69173.1 QOIP01000004 RLU23508.1 KR822176 ALL97694.1 GEZM01034722 JAV83649.1 JX023531 AGE12616.1 LC194140 BAX36479.1 LC194144 BAX36483.1 APGK01013717 KB739085 ENN82392.1 APGK01019652 KB740115 ENN81333.1 KQ434899 KZC10938.1 KB632314 ERL92210.1 KQ976424 KYM88476.1 ADTU01013055 GBYB01013323 JAG83090.1 GBYB01013321 GBYB01013322 JAG83088.1 JAG83089.1 GL887888 EGI69620.1 GECZ01020941 JAS48828.1 GBGD01000076 JAC88813.1 KQ981153 KYN08971.1 JX679512 AFX89025.1 MG225418 AVK59429.1 MG266019 AUM60046.1 KQ981993 KYN30787.1 NNAY01001170 OXU24897.1 DS232090 EDS34500.1 GFTR01008759 JAW07667.1 KU053335 APF46959.1 GEDC01010135 JAS27163.1 GBBI01002022 JAC16690.1 GFDL01010009 JAV25036.1 GFDL01010095 JAV24950.1 LBMM01001117 KMQ96898.1 LC194137 BAW35366.1 KF740508 AIC07485.1 JXUM01089892 KQ563803 KXJ73264.1 KF986375 AIY24625.1 GL451420 EFN79336.1 GGFK01004682 MBW38003.1 GGFK01004697 MBW38018.1 GFDF01002805 JAV11279.1 CH477406 EAT41563.1 JF819823 AEX31249.1 AY713296 AAW48725.1 GANO01003802 JAB56069.1 JXUM01035324 JXUM01035325 JXUM01035326 KQ561055 KXJ79846.1 LC194143 BAX36482.1 JF819824 AEX31250.1 JF819820 JF819821 AEX31247.1 AEX31248.1 ADMH02001044 ETN64340.1 GBHO01016669 JAG26935.1 GDHC01018418 GDHC01005150 JAQ00211.1 JAQ13479.1 GBRD01017340 JAG48487.1 GBHO01016667 JAG26937.1 KK854024 PTY09636.1 LNIX01000017 OXA45600.1 AXCN02001041 GGMR01015856 MBY28475.1 GGMR01002271 MBY14890.1
RVE43638.1 KF717092 AHC98017.1 KQ459037 KPJ04295.1 MG225431 AVK59442.1 KZ150192 PZC72388.1 AGBW02013730 OWR42904.1 ODYU01008375 SOQ51911.1 KQ459833 KPJ19861.1 KP036490 AJF15703.1 LJIG01009215 KRT83525.1 MG225447 AVK59458.1 HE647851 CCF23094.1 KZ288338 PBC27815.1 KP230552 AKQ00041.1 KQ971312 EEZ99277.1 GEZM01034249 JAV83855.1 MH170281 AYE54589.1 KP230553 AKQ00042.1 KQ435851 KOX70787.1 GEZM01034726 GEZM01034725 JAV83643.1 KT191018 AMN92163.1 GEZM01034724 JAV83646.1 JQ037907 AFK73581.1 DS235131 EEB12371.1 GEZM01034723 JAV83648.1 GECU01025228 JAS82478.1 GECU01024018 JAS83688.1 NEVH01022647 PNF18316.1 GL443213 EFN62420.1 KQ982548 KYQ55277.1 KK107054 EZA61552.1 GECZ01000596 JAS69173.1 QOIP01000004 RLU23508.1 KR822176 ALL97694.1 GEZM01034722 JAV83649.1 JX023531 AGE12616.1 LC194140 BAX36479.1 LC194144 BAX36483.1 APGK01013717 KB739085 ENN82392.1 APGK01019652 KB740115 ENN81333.1 KQ434899 KZC10938.1 KB632314 ERL92210.1 KQ976424 KYM88476.1 ADTU01013055 GBYB01013323 JAG83090.1 GBYB01013321 GBYB01013322 JAG83088.1 JAG83089.1 GL887888 EGI69620.1 GECZ01020941 JAS48828.1 GBGD01000076 JAC88813.1 KQ981153 KYN08971.1 JX679512 AFX89025.1 MG225418 AVK59429.1 MG266019 AUM60046.1 KQ981993 KYN30787.1 NNAY01001170 OXU24897.1 DS232090 EDS34500.1 GFTR01008759 JAW07667.1 KU053335 APF46959.1 GEDC01010135 JAS27163.1 GBBI01002022 JAC16690.1 GFDL01010009 JAV25036.1 GFDL01010095 JAV24950.1 LBMM01001117 KMQ96898.1 LC194137 BAW35366.1 KF740508 AIC07485.1 JXUM01089892 KQ563803 KXJ73264.1 KF986375 AIY24625.1 GL451420 EFN79336.1 GGFK01004682 MBW38003.1 GGFK01004697 MBW38018.1 GFDF01002805 JAV11279.1 CH477406 EAT41563.1 JF819823 AEX31249.1 AY713296 AAW48725.1 GANO01003802 JAB56069.1 JXUM01035324 JXUM01035325 JXUM01035326 KQ561055 KXJ79846.1 LC194143 BAX36482.1 JF819824 AEX31250.1 JF819820 JF819821 AEX31247.1 AEX31248.1 ADMH02001044 ETN64340.1 GBHO01016669 JAG26935.1 GDHC01018418 GDHC01005150 JAQ00211.1 JAQ13479.1 GBRD01017340 JAG48487.1 GBHO01016667 JAG26937.1 KK854024 PTY09636.1 LNIX01000017 OXA45600.1 AXCN02001041 GGMR01015856 MBY28475.1 GGMR01002271 MBY14890.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000192223 UP000242457 UP000007266 UP000053105 UP000005203 UP000009046 UP000235965 UP000000311 UP000075809 UP000053097 UP000279307 UP000019118 UP000076502 UP000030742 UP000078540 UP000005205 UP000007755 UP000078492 UP000002358 UP000078541 UP000215335 UP000002320 UP000036403 UP000069940 UP000249989 UP000008237 UP000008820 UP000069272 UP000000673 UP000075885 UP000198287 UP000075886 UP000076408
UP000192223 UP000242457 UP000007266 UP000053105 UP000005203 UP000009046 UP000235965 UP000000311 UP000075809 UP000053097 UP000279307 UP000019118 UP000076502 UP000030742 UP000078540 UP000005205 UP000007755 UP000078492 UP000002358 UP000078541 UP000215335 UP000002320 UP000036403 UP000069940 UP000249989 UP000008237 UP000008820 UP000069272 UP000000673 UP000075885 UP000198287 UP000075886 UP000076408
Pfam
Interpro
IPR000999
RNase_III_dom
+ More
IPR014720 dsRBD_dom
IPR011907 RNase_III
IPR036389 RNase_III_sf
IPR005034 Dicer_dimerisation_dom
IPR038248 Dicer_dimer_sf
IPR006935 Helicase/UvrB_N
IPR027417 P-loop_NTPase
IPR003100 PAZ_dom
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR036085 PAZ_dom_sf
IPR011545 DEAD/DEAH_box_helicase_dom
IPR018247 EF_Hand_1_Ca_BS
IPR014720 dsRBD_dom
IPR011907 RNase_III
IPR036389 RNase_III_sf
IPR005034 Dicer_dimerisation_dom
IPR038248 Dicer_dimer_sf
IPR006935 Helicase/UvrB_N
IPR027417 P-loop_NTPase
IPR003100 PAZ_dom
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR036085 PAZ_dom_sf
IPR011545 DEAD/DEAH_box_helicase_dom
IPR018247 EF_Hand_1_Ca_BS
Gene 3D
ProteinModelPortal
H9JPT8
D7UT11
A0A2A4JFH8
A0A3S2LSR5
V9VKP3
A0A0N1IDN4
+ More
A0A2P1DM58 A0A2W1BBL1 A0A212EN34 A0A2H1WFS6 A0A194RUP4 A0A1W4X5J6 A0A0B5G8Z8 A0A0T6B8D4 A0A2P1DM78 K0JC91 A0A2A3E7Y1 A0A0H4SZH3 D6WA66 A0A1Y1MHP6 A0A386QX51 A0A0H4T2N5 A0A0M8ZTS6 A0A088ABX2 A0A1Y1MD92 A0A140H4W2 A0A1Y1MIC1 K9LIV6 E0VG65 A0A1Y1MF64 A0A1B6I6C5 A0A1B6I9V0 A0A2J7PPT2 E2AVZ8 A0A151X4G5 A0A026WZU4 A0A1B6H3B9 A0A3L8DUT7 A0A0S1RSS9 A0A1Y1MCU9 L7Z6F8 A0A1W7HGU9 A0A1W7HGU4 N6UUA9 N6TTD5 A0A154PHY9 U4UQA5 A0A195BR89 A0A158NDW6 A0A0C9QFQ7 A0A0C9RZ40 F4W7V0 A0A1B6FF57 A0A069DXQ4 A0A195D7X2 K7YZ50 A0A2P1DM48 A0A2I6PIZ5 K7J5H5 A0A195ESA7 A0A232F391 B0WTP2 A0A224X595 A0A1L3A5U6 A0A1B6DNE2 A0A023F657 A0A1Q3FBT5 A0A1Q3FBL4 A0A0J7L2K8 A0A1L7NZN0 A0A060H2K4 A0A182GUS7 A0A0A7DVT8 E2BY57 A0A2M4AB33 A0A2M4ABM6 A0A1L8DXZ8 Q174T8 H2BNN0 Q49LL4 W4VRF8 A0A182FU44 A0A182HE85 A0A1W7HGU5 H2BNN1 H2BNM7 W5JN74 A0A0A9YBL6 A0A146M220 A0A0K8S5D2 A0A0A9Y1K7 A0A182PBR4 A0A2R7VQG7 A0A226DKF2 A0A182PZN5 A0A182Y1T1 A0A2S2PGD2 A0A2S2NCI3
A0A2P1DM58 A0A2W1BBL1 A0A212EN34 A0A2H1WFS6 A0A194RUP4 A0A1W4X5J6 A0A0B5G8Z8 A0A0T6B8D4 A0A2P1DM78 K0JC91 A0A2A3E7Y1 A0A0H4SZH3 D6WA66 A0A1Y1MHP6 A0A386QX51 A0A0H4T2N5 A0A0M8ZTS6 A0A088ABX2 A0A1Y1MD92 A0A140H4W2 A0A1Y1MIC1 K9LIV6 E0VG65 A0A1Y1MF64 A0A1B6I6C5 A0A1B6I9V0 A0A2J7PPT2 E2AVZ8 A0A151X4G5 A0A026WZU4 A0A1B6H3B9 A0A3L8DUT7 A0A0S1RSS9 A0A1Y1MCU9 L7Z6F8 A0A1W7HGU9 A0A1W7HGU4 N6UUA9 N6TTD5 A0A154PHY9 U4UQA5 A0A195BR89 A0A158NDW6 A0A0C9QFQ7 A0A0C9RZ40 F4W7V0 A0A1B6FF57 A0A069DXQ4 A0A195D7X2 K7YZ50 A0A2P1DM48 A0A2I6PIZ5 K7J5H5 A0A195ESA7 A0A232F391 B0WTP2 A0A224X595 A0A1L3A5U6 A0A1B6DNE2 A0A023F657 A0A1Q3FBT5 A0A1Q3FBL4 A0A0J7L2K8 A0A1L7NZN0 A0A060H2K4 A0A182GUS7 A0A0A7DVT8 E2BY57 A0A2M4AB33 A0A2M4ABM6 A0A1L8DXZ8 Q174T8 H2BNN0 Q49LL4 W4VRF8 A0A182FU44 A0A182HE85 A0A1W7HGU5 H2BNN1 H2BNM7 W5JN74 A0A0A9YBL6 A0A146M220 A0A0K8S5D2 A0A0A9Y1K7 A0A182PBR4 A0A2R7VQG7 A0A226DKF2 A0A182PZN5 A0A182Y1T1 A0A2S2PGD2 A0A2S2NCI3
PDB
6BUA
E-value=5.03093e-50,
Score=499
Ontologies
GO
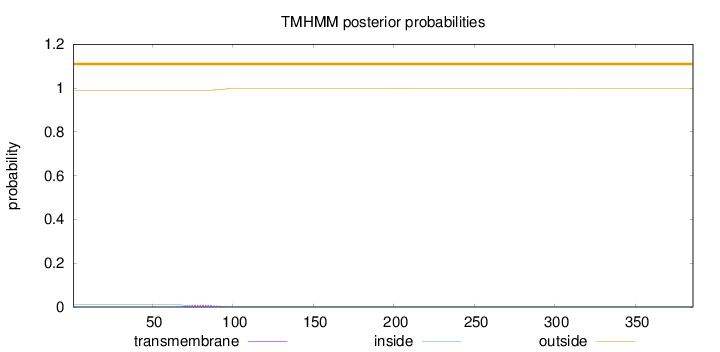
Topology
Length:
386
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2096
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01097
outside
1 - 386
Population Genetic Test Statistics
Pi
315.687643
Theta
180.288861
Tajima's D
2.272671
CLR
0.209913
CSRT
0.921653917304135
Interpretation
Uncertain
