Gene
KWMTBOMO13795
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.207
Sequence
CDS
ATGGATAGGGAAATTCTGTCCATTGACAGTCTGGCTCATAGTGCGAAGGAGAAGGCTGACCTTCTGGTCAAGCTCTTTGCCTCATACTCGACTTTGGATGACGGAGGAGCCACGCCGCCCAACATCTCCCGGTGTGACAGCTCCATGCCTGAGATTCGCATCACACAGCGTGCGGTCAGGCAGGAGCTTCGGCTTCTTGATGTCCATAAGTCGAGTGGGCCAGATTGCATCCCCGCAGTGGTTCTAAAAACATGCGCCCCTGAATTGACACCCGCGCTAACGCGTCTATATCGCCTCTCGTACAATACCAACAGGGTTCCGTCTTCATGGAAGACTGCTCATGTCCACCCTATCCCCAAGAAGGGTGACCGGTCGGACCCCTCGAATTACAGACCTATCGCGATAACTTCCTTGCTTTCCAAGGTGATGGAGCGAATCATAAACATCCAACTTCTTAAGTATCTTGAGGATCGCCAGCTGATCAGTGACCGGCAGTACGGTTTCCGTCATGGTCGCTCAGCTGGCAATCTTCTTGTATACCTCACACATAAATGGGCTGAAGCCTTGGAAAGCAAGGGCGAGGCTCTCGCTGTGAGCCTTGACATCGCGAAGGCCTTCGACAGGGTCTGGCATAGGGCACTTCTATCGAAGCTTCCAACATACGGGATCCCTGAGGGACTCTGCAAGTGGATCGCTAGCTTTTTGGATGGACGGAGCATCGCAGTTGTCGTCGACGGCTGCTGTTCCGATACCATGACCATCAACGCTGGTGTTCCACAAGGTTCGGTGCTCTCCCCCACGCTTTTCATACTGCATATCAATGACATGCTATCTATTGATGGCATGCATTGCTATGCGGATGACAGCACGGGGGATGCGCGATATATCGGCCATCAGAGTCTCTCTCGAAGCGTTGTACACGAGAGGCGATTAAAACTTGTGTCTAAAGTGGAGAACTCTCTGGGGCGGGTCTCCAAGTGGGGTGAACTGAATTCAGTTCAGTTCAACCCATTGAAGACACAGGTTTGCGCGTTCACTGCGAAGAAAGACCCTTTTGTTATGGCGCCGGAATTTCAAGGAGTGTCCCTGCAGCTTTCCGCGAGTATCGGGATTCTGGGGGTCGATATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGTAAAGCCAAATTGGCCTCTAAAATGTTGGAAGTTCTCAACATAGCGAAGCGGTATTTCACGCCTGGACAGAGACTTGCGCTTTATAAAGCGCAGGTCCGACCTCGCGTGGAGTACTGCTCTCACCTCTGGGCCGGGGCTCCCAAATACCAGCTACTTCCATTTGACTCCATACAGAAACGGGCTGTTCGGATGGTCGATAGTCCTGCTCTCGCGGATCGCTTGGAACCTCTGGGTCTATGGAGGGACTTCGGTTCTCTCTGTGTTTTATGCCGCATGGTCCATGGGGAATGCTCTGAGGAATTATTCGAGATGATTCCCTCATCTCCTTTTTATCATCGCACCTCCCGCCATCGGAGCAGAGTTCATCCATATTATCTGGAACCGCTGCGTTCATCGACAGTGCGTTTCCAAAGATCTTTTTTGCCACGTACCATCCGGCTTTGGAATGAACTCCCCTCCACGGTGTTTTCCGAGCGCTATGACATGTCCTTCTTCAAACGAGGCTTGCGGAGAGTTCTTTATGGTAGGCAGCGGCTTGGCTCTGCCCCTGGCATTGCTGACGTCCATGAGCGACGGTGA
Protein
MDREILSIDSLAHSAKEKADLLVKLFASYSTLDDGGATPPNISRCDSSMPEIRITQRAVRQELRLLDVHKSSGPDCIPAVVLKTCAPELTPALTRLYRLSYNTNRVPSSWKTAHVHPIPKKGDRSDPSNYRPIAITSLLSKVMERIINIQLLKYLEDRQLISDRQYGFRHGRSAGNLLVYLTHKWAEALESKGEALAVSLDIAKAFDRVWHRALLSKLPTYGIPEGLCKWIASFLDGRSIAVVVDGCCSDTMTINAGVPQGSVLSPTLFILHINDMLSIDGMHCYADDSTGDARYIGHQSLSRSVVHERRLKLVSKVENSLGRVSKWGELNSVQFNPLKTQVCAFTAKKDPFVMAPEFQGVSLQLSASIGILGVDISSDVQFRSHLEGKAKLASKMLEVLNIAKRYFTPGQRLALYKAQVRPRVEYCSHLWAGAPKYQLLPFDSIQKRAVRMVDSPALADRLEPLGLWRDFGSLCVLCRMVHGECSEELFEMIPSSPFYHRTSRHRSRVHPYYLEPLRSSTVRFQRSFLPRTIRLWNELPSTVFSERYDMSFFKRGLRRVLYGRQRLGSAPGIADVHERR
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=5.93938e-05,
Score=112
Ontologies
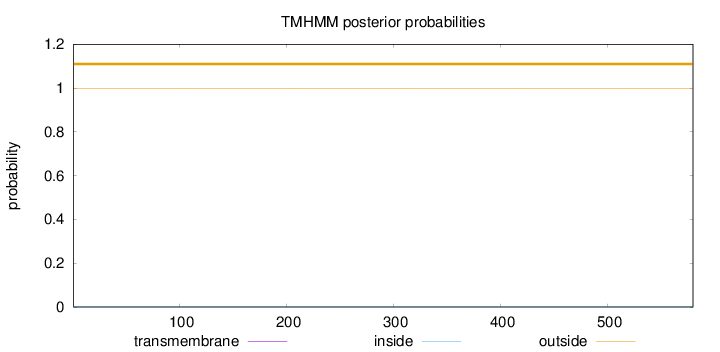
Topology
Length:
580
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00524
Exp number, first 60 AAs:
0.00061
Total prob of N-in:
0.00088
outside
1 - 580
Population Genetic Test Statistics
Pi
31.462811
Theta
45.088774
Tajima's D
1.327759
CLR
0
CSRT
0.753362331883406
Interpretation
Uncertain
