Gene
KWMTBOMO13788
Pre Gene Modal
BGIBMGA011471
Annotation
PREDICTED:_protein_couch_potato_[Papilio_machaon]
Full name
Protein couch potato
Location in the cell
Nuclear Reliability : 1.986
Sequence
CDS
ATGGATGCGAAGCCACGCGAGCTGTATCTACTGTTTCGAGCATATGAGGGCTACGAAGGTTCCCTTCTGAAGGTTACAAGTAAGAACGGGAAGACGGCTTCGCCGGTTGGCTTCGTAACGTTCCACACTCGTGCCGGTGCCGAAGCGGCTAAGCAGGATTTACAGCAGGGCGTCCGGTTCGACCCCGACATGCCGCAAACGATTCGGTTGGAGTTCGCTAAAAGCAACACGAAGGTCAGCAAGCCCAAGCCGGCAGTTGCCACAGCCGCGCCAGCCGCACACCCCGCATTGATGCACCCACTCACTGGACATTTAGCGAGTCCGTTTTTCCCTGGCGGTGGAGCTGAGCTGTGGCACCACCCGCTGGCGTATGGCGGTGAGCTGCCCGGACTCTCGCACGGCCTCGTGCATCCCGCCATCCACCCACAGATGGCGCCCGTGCGTTCCTACCTCTGA
Protein
MDAKPRELYLLFRAYEGYEGSLLKVTSKNGKTASPVGFVTFHTRAGAEAAKQDLQQGVRFDPDMPQTIRLEFAKSNTKVSKPKPAVATAAPAAHPALMHPLTGHLASPFFPGGGAELWHHPLAYGGELPGLSHGLVHPAIHPQMAPVRSYL
Summary
Description
May play a role in the development or function of the peripheral nervous system by regulating the processing of nervous system-specific transcripts.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Nucleus
Reference proteome
Repeat
RNA-binding
Feature
chain Protein couch potato
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A212ESR7
A0A2W1BCU7
A0A194RQV0
A0A0N1IN00
A0A2A4JS44
A0A2H1VS13
+ More
V9IAT2 A0A1Y1KU68 A0A0C9QEX1 A0A1B6CUQ7 H9JPL6 W8C3H2 A0A2R7WLM3 A0A3B0KBD6 A0A0A1WTP2 F6KSN4 A0A034WDV7 A0A0K8UF63 A0A0L7QNA5 T1PMG4 A0A2A3ELY8 V9I9G9 B0WY46 A0A0K8TM31 A0A1A9VPE3 A0A139WND8 A0A087ZZH7 A0A139WNH4 A0A3L8E5N8 A0A158P123 A0A1B6DWR1 B4JUU1 B4QV28 A0A0R3NK64 E2A5V1 Q01617-6 A0A1J1J118 U5EP95 A0A1J1J574 A0A2R7WMN8 A0A1I8JVI8 A0A1I8JT68 A0A182RIL3 A0A182VZJ7 A0A1B6EB19 A0A1I8MWM2 A0A0Q9WFV4 A0A026VUN2 B4KCE7 A0A2M4A6Q6 A0A1I8QEU9 A0A0M4EM63 A0A2M3YZA3 B4NB66 B4PKJ9 A0A2M4BS68 A0A1Q3FU21 A0A0T6B995 A0A1W4U7H4 A0A1B6HGP7 A0A1B6L6A2 A0A232F5X2 A0A1B6EWC1 A0A2M4BNL8 A0A2M4A3H6 A0A2M4A3R1 A0A2M3ZFQ4 T1E2W3 T1DCY4 A0A2S2PU73 E0VPJ7 A0A069DYB6 B4GLJ4 A0A2M4CR49 A0A0B4KGD8 Q01617 A0A2M4CR81 A0A2M4CSG2 A0A0V0GBH5 B4IBB6 A0A1Q3FX61 A0A0B4KHB6 A0A0B4KGV8 A0A2M3YY15 A0A0K8SMI8 A0A0B4KHP4 A0A0K8T8U7 A0A2H8TFU1 A0A182NRW2 A0A182PSL3 F5HMI6 A0A182MP53 B3P0E7 A0A023EUP3 A0A1B6J8D7 A0A2S2P051 J9JJ34 F5HMI5 A0A336LJQ0 A0A336LL31
V9IAT2 A0A1Y1KU68 A0A0C9QEX1 A0A1B6CUQ7 H9JPL6 W8C3H2 A0A2R7WLM3 A0A3B0KBD6 A0A0A1WTP2 F6KSN4 A0A034WDV7 A0A0K8UF63 A0A0L7QNA5 T1PMG4 A0A2A3ELY8 V9I9G9 B0WY46 A0A0K8TM31 A0A1A9VPE3 A0A139WND8 A0A087ZZH7 A0A139WNH4 A0A3L8E5N8 A0A158P123 A0A1B6DWR1 B4JUU1 B4QV28 A0A0R3NK64 E2A5V1 Q01617-6 A0A1J1J118 U5EP95 A0A1J1J574 A0A2R7WMN8 A0A1I8JVI8 A0A1I8JT68 A0A182RIL3 A0A182VZJ7 A0A1B6EB19 A0A1I8MWM2 A0A0Q9WFV4 A0A026VUN2 B4KCE7 A0A2M4A6Q6 A0A1I8QEU9 A0A0M4EM63 A0A2M3YZA3 B4NB66 B4PKJ9 A0A2M4BS68 A0A1Q3FU21 A0A0T6B995 A0A1W4U7H4 A0A1B6HGP7 A0A1B6L6A2 A0A232F5X2 A0A1B6EWC1 A0A2M4BNL8 A0A2M4A3H6 A0A2M4A3R1 A0A2M3ZFQ4 T1E2W3 T1DCY4 A0A2S2PU73 E0VPJ7 A0A069DYB6 B4GLJ4 A0A2M4CR49 A0A0B4KGD8 Q01617 A0A2M4CR81 A0A2M4CSG2 A0A0V0GBH5 B4IBB6 A0A1Q3FX61 A0A0B4KHB6 A0A0B4KGV8 A0A2M3YY15 A0A0K8SMI8 A0A0B4KHP4 A0A0K8T8U7 A0A2H8TFU1 A0A182NRW2 A0A182PSL3 F5HMI6 A0A182MP53 B3P0E7 A0A023EUP3 A0A1B6J8D7 A0A2S2P051 J9JJ34 F5HMI5 A0A336LJQ0 A0A336LL31
Pubmed
22118469
28756777
26354079
28004739
19121390
24495485
+ More
25830018 21315077 25348373 26369729 18362917 19820115 30249741 21347285 17994087 15632085 20798317 1427076 10731132 12537572 12537569 18852464 25315136 24508170 17550304 28648823 24330624 20566863 26334808 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 24945155
25830018 21315077 25348373 26369729 18362917 19820115 30249741 21347285 17994087 15632085 20798317 1427076 10731132 12537572 12537569 18852464 25315136 24508170 17550304 28648823 24330624 20566863 26334808 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 24945155
EMBL
AGBW02012729
OWR44527.1
KZ150207
PZC72241.1
KQ459833
KPJ19857.1
+ More
KQ459037 KPJ04290.1 NWSH01000777 PCG74293.1 ODYU01004111 SOQ43641.1 JR037410 AEY57757.1 GEZM01073676 GEZM01073675 GEZM01073674 GEZM01073673 JAV64939.1 GBYB01001884 GBYB01001886 JAG71651.1 JAG71653.1 GEDC01020233 GEDC01019186 JAS17065.1 JAS18112.1 BABH01015274 GAMC01010131 JAB96424.1 KK855009 PTY20271.1 OUUW01000007 SPP83429.1 GBXI01011858 JAD02434.1 HQ603004 ADW77182.1 GAKP01005231 GAKP01005229 JAC53721.1 GDHF01027007 GDHF01021518 GDHF01003315 GDHF01001373 JAI25307.1 JAI30796.1 JAI48999.1 JAI50941.1 KQ414855 KOC60110.1 KA649113 AFP63742.1 KZ288212 PBC32813.1 JR037412 AEY57758.1 DS232180 EDS36856.1 GDAI01002181 JAI15422.1 KQ971311 KYB29452.1 KYB29453.1 QOIP01000001 RLU27168.1 ADTU01006086 ADTU01006087 ADTU01006088 ADTU01006089 ADTU01006090 ADTU01006091 ADTU01006092 ADTU01006093 ADTU01006094 ADTU01006095 GEDC01007174 JAS30124.1 CH916374 EDV91261.1 CM000364 EDX12511.1 CM000070 KRT01298.1 GL437061 EFN71193.1 Z14311 Z14312 Z14974 AE014297 AY129455 BT001569 CVRI01000066 CRL06032.1 GANO01003771 JAB56100.1 CRL06033.1 PTY20270.1 APCN01000909 GEDC01002185 JAS35113.1 CH940650 KRF83106.1 KK107921 EZA47206.1 CH933806 EDW13756.2 GGFK01003110 MBW36431.1 CP012526 ALC46502.1 GGFM01000842 MBW21593.1 CH964232 EDW81030.2 CM000160 EDW95838.2 GGFJ01006786 MBW55927.1 GFDL01004122 JAV30923.1 LJIG01009029 KRT83898.1 GECU01033812 GECU01013920 JAS73894.1 JAS93786.1 GEBQ01020781 JAT19196.1 NNAY01000952 OXU25757.1 GECZ01027494 GECZ01018905 GECZ01000725 JAS42275.1 JAS50864.1 JAS69044.1 GGFJ01005538 MBW54679.1 GGFK01001980 MBW35301.1 GGFK01001957 MBW35278.1 GGFM01006537 MBW27288.1 GALA01000920 JAA93932.1 GALA01001647 JAA93205.1 GGMR01020306 MBY32925.1 DS235366 EEB15303.1 GBGD01002515 JAC86374.1 CH479185 EDW38418.1 GGFL01003628 MBW67806.1 AGB96071.1 AGB96072.1 GGFL01003611 MBW67789.1 GGFL01003610 MBW67788.1 GECL01000698 JAP05426.1 CH480827 EDW44674.1 GFDL01002845 JAV32200.1 AGB96070.1 AGB96068.1 GGFM01000399 MBW21150.1 GBRD01011346 GBRD01011345 JAG54478.1 AGB96069.1 GBRD01010256 GBRD01003867 JAG61954.1 GFXV01001140 MBW12945.1 AAAB01008987 EGK97508.1 AXCM01000938 AXCM01000939 CH954181 EDV48662.1 GAPW01001419 JAC12179.1 GECU01012257 JAS95449.1 GGMR01010196 MBY22815.1 ABLF02031731 EGK97506.1 UFQT01000038 SSX18414.1 SSX18415.1
KQ459037 KPJ04290.1 NWSH01000777 PCG74293.1 ODYU01004111 SOQ43641.1 JR037410 AEY57757.1 GEZM01073676 GEZM01073675 GEZM01073674 GEZM01073673 JAV64939.1 GBYB01001884 GBYB01001886 JAG71651.1 JAG71653.1 GEDC01020233 GEDC01019186 JAS17065.1 JAS18112.1 BABH01015274 GAMC01010131 JAB96424.1 KK855009 PTY20271.1 OUUW01000007 SPP83429.1 GBXI01011858 JAD02434.1 HQ603004 ADW77182.1 GAKP01005231 GAKP01005229 JAC53721.1 GDHF01027007 GDHF01021518 GDHF01003315 GDHF01001373 JAI25307.1 JAI30796.1 JAI48999.1 JAI50941.1 KQ414855 KOC60110.1 KA649113 AFP63742.1 KZ288212 PBC32813.1 JR037412 AEY57758.1 DS232180 EDS36856.1 GDAI01002181 JAI15422.1 KQ971311 KYB29452.1 KYB29453.1 QOIP01000001 RLU27168.1 ADTU01006086 ADTU01006087 ADTU01006088 ADTU01006089 ADTU01006090 ADTU01006091 ADTU01006092 ADTU01006093 ADTU01006094 ADTU01006095 GEDC01007174 JAS30124.1 CH916374 EDV91261.1 CM000364 EDX12511.1 CM000070 KRT01298.1 GL437061 EFN71193.1 Z14311 Z14312 Z14974 AE014297 AY129455 BT001569 CVRI01000066 CRL06032.1 GANO01003771 JAB56100.1 CRL06033.1 PTY20270.1 APCN01000909 GEDC01002185 JAS35113.1 CH940650 KRF83106.1 KK107921 EZA47206.1 CH933806 EDW13756.2 GGFK01003110 MBW36431.1 CP012526 ALC46502.1 GGFM01000842 MBW21593.1 CH964232 EDW81030.2 CM000160 EDW95838.2 GGFJ01006786 MBW55927.1 GFDL01004122 JAV30923.1 LJIG01009029 KRT83898.1 GECU01033812 GECU01013920 JAS73894.1 JAS93786.1 GEBQ01020781 JAT19196.1 NNAY01000952 OXU25757.1 GECZ01027494 GECZ01018905 GECZ01000725 JAS42275.1 JAS50864.1 JAS69044.1 GGFJ01005538 MBW54679.1 GGFK01001980 MBW35301.1 GGFK01001957 MBW35278.1 GGFM01006537 MBW27288.1 GALA01000920 JAA93932.1 GALA01001647 JAA93205.1 GGMR01020306 MBY32925.1 DS235366 EEB15303.1 GBGD01002515 JAC86374.1 CH479185 EDW38418.1 GGFL01003628 MBW67806.1 AGB96071.1 AGB96072.1 GGFL01003611 MBW67789.1 GGFL01003610 MBW67788.1 GECL01000698 JAP05426.1 CH480827 EDW44674.1 GFDL01002845 JAV32200.1 AGB96070.1 AGB96068.1 GGFM01000399 MBW21150.1 GBRD01011346 GBRD01011345 JAG54478.1 AGB96069.1 GBRD01010256 GBRD01003867 JAG61954.1 GFXV01001140 MBW12945.1 AAAB01008987 EGK97508.1 AXCM01000938 AXCM01000939 CH954181 EDV48662.1 GAPW01001419 JAC12179.1 GECU01012257 JAS95449.1 GGMR01010196 MBY22815.1 ABLF02031731 EGK97506.1 UFQT01000038 SSX18414.1 SSX18415.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000005204
UP000268350
+ More
UP000053825 UP000242457 UP000002320 UP000078200 UP000007266 UP000005203 UP000279307 UP000005205 UP000001070 UP000000304 UP000001819 UP000000311 UP000000803 UP000183832 UP000076407 UP000075840 UP000075900 UP000075920 UP000095301 UP000008792 UP000053097 UP000009192 UP000095300 UP000092553 UP000007798 UP000002282 UP000192221 UP000215335 UP000009046 UP000008744 UP000001292 UP000075884 UP000075885 UP000007062 UP000075883 UP000008711 UP000007819
UP000053825 UP000242457 UP000002320 UP000078200 UP000007266 UP000005203 UP000279307 UP000005205 UP000001070 UP000000304 UP000001819 UP000000311 UP000000803 UP000183832 UP000076407 UP000075840 UP000075900 UP000075920 UP000095301 UP000008792 UP000053097 UP000009192 UP000095300 UP000092553 UP000007798 UP000002282 UP000192221 UP000215335 UP000009046 UP000008744 UP000001292 UP000075884 UP000075885 UP000007062 UP000075883 UP000008711 UP000007819
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
CDD
ProteinModelPortal
A0A212ESR7
A0A2W1BCU7
A0A194RQV0
A0A0N1IN00
A0A2A4JS44
A0A2H1VS13
+ More
V9IAT2 A0A1Y1KU68 A0A0C9QEX1 A0A1B6CUQ7 H9JPL6 W8C3H2 A0A2R7WLM3 A0A3B0KBD6 A0A0A1WTP2 F6KSN4 A0A034WDV7 A0A0K8UF63 A0A0L7QNA5 T1PMG4 A0A2A3ELY8 V9I9G9 B0WY46 A0A0K8TM31 A0A1A9VPE3 A0A139WND8 A0A087ZZH7 A0A139WNH4 A0A3L8E5N8 A0A158P123 A0A1B6DWR1 B4JUU1 B4QV28 A0A0R3NK64 E2A5V1 Q01617-6 A0A1J1J118 U5EP95 A0A1J1J574 A0A2R7WMN8 A0A1I8JVI8 A0A1I8JT68 A0A182RIL3 A0A182VZJ7 A0A1B6EB19 A0A1I8MWM2 A0A0Q9WFV4 A0A026VUN2 B4KCE7 A0A2M4A6Q6 A0A1I8QEU9 A0A0M4EM63 A0A2M3YZA3 B4NB66 B4PKJ9 A0A2M4BS68 A0A1Q3FU21 A0A0T6B995 A0A1W4U7H4 A0A1B6HGP7 A0A1B6L6A2 A0A232F5X2 A0A1B6EWC1 A0A2M4BNL8 A0A2M4A3H6 A0A2M4A3R1 A0A2M3ZFQ4 T1E2W3 T1DCY4 A0A2S2PU73 E0VPJ7 A0A069DYB6 B4GLJ4 A0A2M4CR49 A0A0B4KGD8 Q01617 A0A2M4CR81 A0A2M4CSG2 A0A0V0GBH5 B4IBB6 A0A1Q3FX61 A0A0B4KHB6 A0A0B4KGV8 A0A2M3YY15 A0A0K8SMI8 A0A0B4KHP4 A0A0K8T8U7 A0A2H8TFU1 A0A182NRW2 A0A182PSL3 F5HMI6 A0A182MP53 B3P0E7 A0A023EUP3 A0A1B6J8D7 A0A2S2P051 J9JJ34 F5HMI5 A0A336LJQ0 A0A336LL31
V9IAT2 A0A1Y1KU68 A0A0C9QEX1 A0A1B6CUQ7 H9JPL6 W8C3H2 A0A2R7WLM3 A0A3B0KBD6 A0A0A1WTP2 F6KSN4 A0A034WDV7 A0A0K8UF63 A0A0L7QNA5 T1PMG4 A0A2A3ELY8 V9I9G9 B0WY46 A0A0K8TM31 A0A1A9VPE3 A0A139WND8 A0A087ZZH7 A0A139WNH4 A0A3L8E5N8 A0A158P123 A0A1B6DWR1 B4JUU1 B4QV28 A0A0R3NK64 E2A5V1 Q01617-6 A0A1J1J118 U5EP95 A0A1J1J574 A0A2R7WMN8 A0A1I8JVI8 A0A1I8JT68 A0A182RIL3 A0A182VZJ7 A0A1B6EB19 A0A1I8MWM2 A0A0Q9WFV4 A0A026VUN2 B4KCE7 A0A2M4A6Q6 A0A1I8QEU9 A0A0M4EM63 A0A2M3YZA3 B4NB66 B4PKJ9 A0A2M4BS68 A0A1Q3FU21 A0A0T6B995 A0A1W4U7H4 A0A1B6HGP7 A0A1B6L6A2 A0A232F5X2 A0A1B6EWC1 A0A2M4BNL8 A0A2M4A3H6 A0A2M4A3R1 A0A2M3ZFQ4 T1E2W3 T1DCY4 A0A2S2PU73 E0VPJ7 A0A069DYB6 B4GLJ4 A0A2M4CR49 A0A0B4KGD8 Q01617 A0A2M4CR81 A0A2M4CSG2 A0A0V0GBH5 B4IBB6 A0A1Q3FX61 A0A0B4KHB6 A0A0B4KGV8 A0A2M3YY15 A0A0K8SMI8 A0A0B4KHP4 A0A0K8T8U7 A0A2H8TFU1 A0A182NRW2 A0A182PSL3 F5HMI6 A0A182MP53 B3P0E7 A0A023EUP3 A0A1B6J8D7 A0A2S2P051 J9JJ34 F5HMI5 A0A336LJQ0 A0A336LL31
PDB
5TKZ
E-value=3.25674e-23,
Score=262
Ontologies
GO
Topology
Subcellular location
Nucleus
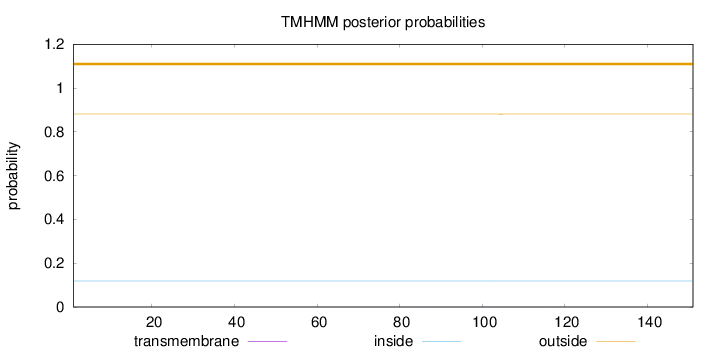
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01364
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.11904
outside
1 - 151
Population Genetic Test Statistics
Pi
258.981463
Theta
212.478523
Tajima's D
0.975927
CLR
0.059732
CSRT
0.654867256637168
Interpretation
Uncertain
