Gene
KWMTBOMO13787 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011540
Annotation
PREDICTED:_uncharacterized_protein_LOC101746356_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.121
Sequence
CDS
ATGGCTTCTAGTGACCCTGAAATTAAAGATGGATTTACACTTCCTGTATGGATGAAGACAAAACCAACAAAAGAATTTGAACCGTTCGCCGGATCGCCGCCCCCACCGGAAGATTCATTCTTCTACATACGCTACCCTAAATCGGATATATTATTTGGTAAACCCACGAATAGTCAAGACGCGCTTGTGAACGAAACGAAGGAAGTGACCGCTGCCTCGGCATTCAACGTCGTCAAGGAGAAAGACTGGTCGAAACACACCAAGCCGTGCCAAGATGTGCTCCACGGCATCGCGCCAATCAACACCGCAGTGAACTCTGTTACGAAACCGAAGCCAGCCGGAACGGATGACGGCTTCAAAGGCGAAGGAGCGGCGTGTGGTACTTCTGTAGTGAAGGTCGCACATCAGATGATATCTTCTCGGTCTCCGCGCGGGTTCACGGTGGCGTCGCCGTCTGAGGACGCCACCGCTCCGGACCCTCAGCACTATAATCTGATGGCGCGTCGAGGCAGCAAGAGTCTCCCCGCCACGCCGGCACACTCGCCCCATTCCAGTCCCACCACCAGAAGGAAGAAGAATGGCAACCGTTACTTCACATCACCGTTCGAGCCAACAGAAGACCCGTCTAACCGATCGTGGTTGTCGATAGCTCTGCTTGGCTTCAAGAAGGAGCTTGCCACTTCAACTTCCACGTTAGCTGAAGAAGACTTGTTGGAACTGAGATCGCAAGCGGGCAGCTTGGCGGAGTCAGTGGAGAGCCTCGGTCCGGCTCCGAAGAACAAAGCACCCGAGAATCCCCACCAGCAGCACGCGTCTCCAACGAAAACAAAACCGAATCATTCCTTCCTCCCGAAACCATCTGAATTGCGCGAAATGAACTTCTGGTCGCCAACGTCAATGTGA
Protein
MASSDPEIKDGFTLPVWMKTKPTKEFEPFAGSPPPPEDSFFYIRYPKSDILFGKPTNSQDALVNETKEVTAASAFNVVKEKDWSKHTKPCQDVLHGIAPINTAVNSVTKPKPAGTDDGFKGEGAACGTSVVKVAHQMISSRSPRGFTVASPSEDATAPDPQHYNLMARRGSKSLPATPAHSPHSSPTTRRKKNGNRYFTSPFEPTEDPSNRSWLSIALLGFKKELATSTSTLAEEDLLELRSQAGSLAESVESLGPAPKNKAPENPHQQHASPTKTKPNHSFLPKPSELREMNFWSPTSM
Summary
Uniprot
A0A1E1W553
A0A0N1PGB8
A0A194RRC1
A0A2A4JQU3
S4PEE6
A0A2H1VWY7
+ More
A0A2W1B973 A0A1E1WIS1 A0A3S2NMM7 A0A212FGT2 A0A0L7KMM7 D6W985 A0A0K8TPB2 A0A1Y1LU05 A0A1B0CJM5 V5G2A0 A0A0J7KJ72 E2A8P2 A0A1L8DD17 A0A151XGL3 A0A195F9B9 A0A151ICY9 E2BNS5 A0A151ISP1 A0A1Y1LSC3 A0A195BNM8 A0A158N9P4 A0A026WEB3 A0A067R553 A0A1W4WZ44 V9IEA9 A0A1W4X9V6 A0A2S2QVR9 A0A154P245 A0A1B6IGY2 A0A2S2PC82 F4WTT6 A0A1B6C5E6 A0A1B0DQ80 A0A088AFI6 A0A0L7QKT5 A0A2P8YHU5 A0A1B6K9K2 R4WRT9 A0A1B6FPE4 J9KPG4 E0VYB4 U4U4E3 A0A2A3E3Z6 A0A1B6GKB9 A0A1B6GXX4 K7J5S0 A0A232FIN2 A0A2R7WME5 A0A0M9ABG9 A0A310S778 A0A224XGR7 N6T2E0 A0A0V0G7R1 A0A023F7E9 C4WRK3 A0A1B6KS04 A0A1B6FWY1 A0A1B6FJK5 A0A0P4W114 R4FNH2 A0A336MDJ6 A0A336MW06 A0A0A9WPN1
A0A2W1B973 A0A1E1WIS1 A0A3S2NMM7 A0A212FGT2 A0A0L7KMM7 D6W985 A0A0K8TPB2 A0A1Y1LU05 A0A1B0CJM5 V5G2A0 A0A0J7KJ72 E2A8P2 A0A1L8DD17 A0A151XGL3 A0A195F9B9 A0A151ICY9 E2BNS5 A0A151ISP1 A0A1Y1LSC3 A0A195BNM8 A0A158N9P4 A0A026WEB3 A0A067R553 A0A1W4WZ44 V9IEA9 A0A1W4X9V6 A0A2S2QVR9 A0A154P245 A0A1B6IGY2 A0A2S2PC82 F4WTT6 A0A1B6C5E6 A0A1B0DQ80 A0A088AFI6 A0A0L7QKT5 A0A2P8YHU5 A0A1B6K9K2 R4WRT9 A0A1B6FPE4 J9KPG4 E0VYB4 U4U4E3 A0A2A3E3Z6 A0A1B6GKB9 A0A1B6GXX4 K7J5S0 A0A232FIN2 A0A2R7WME5 A0A0M9ABG9 A0A310S778 A0A224XGR7 N6T2E0 A0A0V0G7R1 A0A023F7E9 C4WRK3 A0A1B6KS04 A0A1B6FWY1 A0A1B6FJK5 A0A0P4W114 R4FNH2 A0A336MDJ6 A0A336MW06 A0A0A9WPN1
Pubmed
EMBL
GDQN01010150
GDQN01008948
JAT80904.1
JAT82106.1
KQ459037
KPJ04288.1
+ More
KQ459833 KPJ19855.1 NWSH01000798 PCG74146.1 GAIX01007135 JAA85425.1 ODYU01004934 SOQ45318.1 KZ150207 PZC72239.1 GDQN01009094 GDQN01004145 JAT81960.1 JAT86909.1 RSAL01000242 RVE43628.1 AGBW02008614 OWR52913.1 JTDY01008820 KOB64380.1 KQ971312 EEZ98200.1 GDAI01001376 JAI16227.1 GEZM01052498 JAV74477.1 AJWK01014748 GALX01004326 JAB64140.1 LBMM01006599 KMQ90473.1 GL437663 EFN70121.1 GFDF01009723 JAV04361.1 KQ982169 KYQ59536.1 KQ981727 KYN36649.1 KQ977991 KYM98281.1 GL449462 EFN82658.1 KQ981064 KYN09882.1 GEZM01052499 JAV74476.1 KQ976428 KYM87827.1 ADTU01009726 KK107260 QOIP01000001 EZA54006.1 RLU26448.1 KK852962 KDR13202.1 JR039306 AEY58629.1 GGMS01012648 MBY81851.1 KQ434803 KZC06009.1 GECU01021537 JAS86169.1 GGMR01014444 MBY27063.1 GL888344 EGI62401.1 GEDC01028566 JAS08732.1 AJVK01018926 AJVK01018927 AJVK01018928 KQ414940 KOC59136.1 PYGN01000583 PSN43825.1 GEBQ01031849 JAT08128.1 AK417417 BAN20632.1 GECZ01017701 GECZ01017148 JAS52068.1 JAS52621.1 ABLF02010726 ABLF02010728 DS235845 EEB18370.1 KB631924 ERL87218.1 KZ288422 PBC25976.1 GECZ01006894 JAS62875.1 GECZ01025071 GECZ01002491 GECZ01000849 JAS44698.1 JAS67278.1 JAS68920.1 AAZX01011597 AAZX01013086 NNAY01000159 OXU30443.1 KK855095 PTY20796.1 KQ435710 KOX79739.1 KQ780658 OAD51919.1 GFTR01004739 JAW11687.1 APGK01055063 APGK01055064 KB741259 ENN71703.1 ERL87219.1 GECL01002642 JAP03482.1 GBBI01001542 JAC17170.1 AK339808 BAH70523.1 GEBQ01025759 JAT14218.1 GECZ01018297 GECZ01015110 GECZ01000965 JAS51472.1 JAS54659.1 JAS68804.1 GECZ01019383 GECZ01017000 JAS50386.1 JAS52769.1 GDKW01000005 JAI56590.1 ACPB03021640 GAHY01000573 JAA76937.1 UFQT01000827 SSX27421.1 UFQT01002674 SSX33905.1 GBHO01035156 GBHO01035155 GDHC01013332 GDHC01000931 JAG08448.1 JAG08449.1 JAQ05297.1 JAQ17698.1
KQ459833 KPJ19855.1 NWSH01000798 PCG74146.1 GAIX01007135 JAA85425.1 ODYU01004934 SOQ45318.1 KZ150207 PZC72239.1 GDQN01009094 GDQN01004145 JAT81960.1 JAT86909.1 RSAL01000242 RVE43628.1 AGBW02008614 OWR52913.1 JTDY01008820 KOB64380.1 KQ971312 EEZ98200.1 GDAI01001376 JAI16227.1 GEZM01052498 JAV74477.1 AJWK01014748 GALX01004326 JAB64140.1 LBMM01006599 KMQ90473.1 GL437663 EFN70121.1 GFDF01009723 JAV04361.1 KQ982169 KYQ59536.1 KQ981727 KYN36649.1 KQ977991 KYM98281.1 GL449462 EFN82658.1 KQ981064 KYN09882.1 GEZM01052499 JAV74476.1 KQ976428 KYM87827.1 ADTU01009726 KK107260 QOIP01000001 EZA54006.1 RLU26448.1 KK852962 KDR13202.1 JR039306 AEY58629.1 GGMS01012648 MBY81851.1 KQ434803 KZC06009.1 GECU01021537 JAS86169.1 GGMR01014444 MBY27063.1 GL888344 EGI62401.1 GEDC01028566 JAS08732.1 AJVK01018926 AJVK01018927 AJVK01018928 KQ414940 KOC59136.1 PYGN01000583 PSN43825.1 GEBQ01031849 JAT08128.1 AK417417 BAN20632.1 GECZ01017701 GECZ01017148 JAS52068.1 JAS52621.1 ABLF02010726 ABLF02010728 DS235845 EEB18370.1 KB631924 ERL87218.1 KZ288422 PBC25976.1 GECZ01006894 JAS62875.1 GECZ01025071 GECZ01002491 GECZ01000849 JAS44698.1 JAS67278.1 JAS68920.1 AAZX01011597 AAZX01013086 NNAY01000159 OXU30443.1 KK855095 PTY20796.1 KQ435710 KOX79739.1 KQ780658 OAD51919.1 GFTR01004739 JAW11687.1 APGK01055063 APGK01055064 KB741259 ENN71703.1 ERL87219.1 GECL01002642 JAP03482.1 GBBI01001542 JAC17170.1 AK339808 BAH70523.1 GEBQ01025759 JAT14218.1 GECZ01018297 GECZ01015110 GECZ01000965 JAS51472.1 JAS54659.1 JAS68804.1 GECZ01019383 GECZ01017000 JAS50386.1 JAS52769.1 GDKW01000005 JAI56590.1 ACPB03021640 GAHY01000573 JAA76937.1 UFQT01000827 SSX27421.1 UFQT01002674 SSX33905.1 GBHO01035156 GBHO01035155 GDHC01013332 GDHC01000931 JAG08448.1 JAG08449.1 JAQ05297.1 JAQ17698.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
UP000037510
+ More
UP000007266 UP000092461 UP000036403 UP000000311 UP000075809 UP000078541 UP000078542 UP000008237 UP000078492 UP000078540 UP000005205 UP000053097 UP000279307 UP000027135 UP000192223 UP000076502 UP000007755 UP000092462 UP000005203 UP000053825 UP000245037 UP000007819 UP000009046 UP000030742 UP000242457 UP000002358 UP000215335 UP000053105 UP000019118 UP000015103
UP000007266 UP000092461 UP000036403 UP000000311 UP000075809 UP000078541 UP000078542 UP000008237 UP000078492 UP000078540 UP000005205 UP000053097 UP000279307 UP000027135 UP000192223 UP000076502 UP000007755 UP000092462 UP000005203 UP000053825 UP000245037 UP000007819 UP000009046 UP000030742 UP000242457 UP000002358 UP000215335 UP000053105 UP000019118 UP000015103
Pfam
PF11018 Cuticle_3
Interpro
IPR022727
Cuticle_C1
ProteinModelPortal
A0A1E1W553
A0A0N1PGB8
A0A194RRC1
A0A2A4JQU3
S4PEE6
A0A2H1VWY7
+ More
A0A2W1B973 A0A1E1WIS1 A0A3S2NMM7 A0A212FGT2 A0A0L7KMM7 D6W985 A0A0K8TPB2 A0A1Y1LU05 A0A1B0CJM5 V5G2A0 A0A0J7KJ72 E2A8P2 A0A1L8DD17 A0A151XGL3 A0A195F9B9 A0A151ICY9 E2BNS5 A0A151ISP1 A0A1Y1LSC3 A0A195BNM8 A0A158N9P4 A0A026WEB3 A0A067R553 A0A1W4WZ44 V9IEA9 A0A1W4X9V6 A0A2S2QVR9 A0A154P245 A0A1B6IGY2 A0A2S2PC82 F4WTT6 A0A1B6C5E6 A0A1B0DQ80 A0A088AFI6 A0A0L7QKT5 A0A2P8YHU5 A0A1B6K9K2 R4WRT9 A0A1B6FPE4 J9KPG4 E0VYB4 U4U4E3 A0A2A3E3Z6 A0A1B6GKB9 A0A1B6GXX4 K7J5S0 A0A232FIN2 A0A2R7WME5 A0A0M9ABG9 A0A310S778 A0A224XGR7 N6T2E0 A0A0V0G7R1 A0A023F7E9 C4WRK3 A0A1B6KS04 A0A1B6FWY1 A0A1B6FJK5 A0A0P4W114 R4FNH2 A0A336MDJ6 A0A336MW06 A0A0A9WPN1
A0A2W1B973 A0A1E1WIS1 A0A3S2NMM7 A0A212FGT2 A0A0L7KMM7 D6W985 A0A0K8TPB2 A0A1Y1LU05 A0A1B0CJM5 V5G2A0 A0A0J7KJ72 E2A8P2 A0A1L8DD17 A0A151XGL3 A0A195F9B9 A0A151ICY9 E2BNS5 A0A151ISP1 A0A1Y1LSC3 A0A195BNM8 A0A158N9P4 A0A026WEB3 A0A067R553 A0A1W4WZ44 V9IEA9 A0A1W4X9V6 A0A2S2QVR9 A0A154P245 A0A1B6IGY2 A0A2S2PC82 F4WTT6 A0A1B6C5E6 A0A1B0DQ80 A0A088AFI6 A0A0L7QKT5 A0A2P8YHU5 A0A1B6K9K2 R4WRT9 A0A1B6FPE4 J9KPG4 E0VYB4 U4U4E3 A0A2A3E3Z6 A0A1B6GKB9 A0A1B6GXX4 K7J5S0 A0A232FIN2 A0A2R7WME5 A0A0M9ABG9 A0A310S778 A0A224XGR7 N6T2E0 A0A0V0G7R1 A0A023F7E9 C4WRK3 A0A1B6KS04 A0A1B6FWY1 A0A1B6FJK5 A0A0P4W114 R4FNH2 A0A336MDJ6 A0A336MW06 A0A0A9WPN1
Ontologies
GO
PANTHER
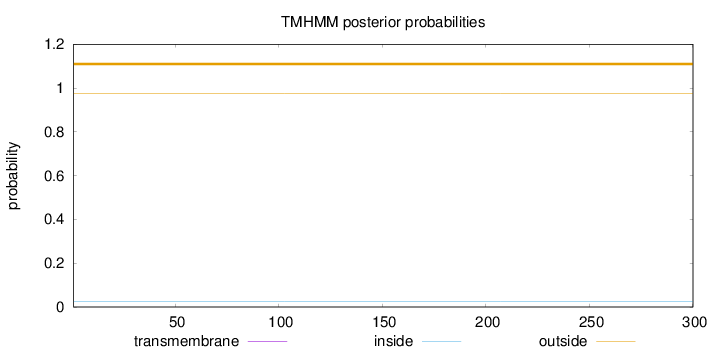
Topology
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02439
outside
1 - 300
Population Genetic Test Statistics
Pi
277.548135
Theta
186.63499
Tajima's D
1.309223
CLR
0.116355
CSRT
0.742462876856157
Interpretation
Uncertain
