Pre Gene Modal
BGIBMGA011537
Annotation
PREDICTED:_UPF0668_protein_C10orf76_homolog_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 3.085
Sequence
CDS
ATGGCGATGAGAAAAAGAAGTGGCTCTGGATCAAAGAGACACTTAAAAGAGAAGGTTGTACAAATTTATGAATCGTTCTTTCGAGGAGAGGACTTAACGACTGATAACCCTACTTTTTGGGATGAATTCTTTCTTCTGAAGCCAAAAATACCACAGCTGGAAGCAGAAATACACAAACTGTCCACAGAACAGCTGATAAACATGAAAGAAAGTATGAATATATTGGTTGAACAGTGTATAGAAATGTTGAGACAAGACCATCATATAAGGTTGCTTTATGCTTTACAAACATTAAGTGCATTACTCATTACTATGTATCAAAGACTTGAACAGGATTCAAACATAAACATCAAAACTATAATGATTGGGCAAGAGAATGCTGATACTAAAATGGTGAAATTATTTGAACAATGCAGTATTATACTATCAGGTGATGTTCATGAGAGCTTAAAGTCAATGGTTCTGAAGCTCTTGCTTATTATAGCAACTGGAATAGAAAATATAGATGAGAATCCTTTTGTGGAGTATTTCATGTCAAACTGTTTGTTTGAGCCATTGATACAGCTGCTTTGTACCACTACAGAAAGGCAACAGCATGGTTATGATGTAGTTATGTTACTGATGTTTTTGATAAACTATAAGAAGCATGAATCAACAAATCCATATGTTGTTAAACTTTCTATATTAGATGATGAGCTGGCTTTGAACGGCTATGGCCAAGTAATAACGACAGCTTTGAATGACTATGTAATGTCAGCATTCGGTGGTATGCAAGGCAACGGGGGTGGAGGTGGTTGGTTGTCATCTCTAACTAGCATGGTTGGGGGTATATTCCTAACAAATGATGAACAACAACCTTTAGCCAGGGGACAAAGGAACAATGGGGCCGAAGAAGGAATGCTCCTGGCTATGTATGAGGCTTCGCATTTAAATCGGAATTTTATGACGACTTTGGCACATTCGTCTTCAACGACGGCGTCGTCAGCGCCACCATCGCCCCCAGCGACGTTACCGCCTCACCAGAGTCCACCGAACCTCGCTCAAATACAAGCCCTGGACAATGATCAACCAACGAATTTACTAGTCACATTCTTTCAGTATTGTTCAATCATAATGGCGGACACCAGGAGCGAAAGTAGTATTAACAAGTGCAGTCTTTGTTTTATCACTTTAACGTGCATCGTGGAGGACCAATTTGCTAATTCAATTATGCACGACCAAAACCTCACTTTTAAGGTACAATTATATAGACTTCCGATGCGTCATAGGAAAATAGTTCTTGAGGAACCACCGTCACAACCGTTGGCGTCTACTTTAATAGATCTTTTAGTCGAGTTCATAATGTGTCACCTGCTGAAGAAGTTCCCCGGCGAGCTGTATAGCCTGTGCGTGGGCGTGCTCCTCCGGCTACTGAGCTACCAGAAGCGGTGCAAGGTTCGTCTGGCGCGAGACTGGCGACCTCTGTGGGCCGCACTCATAGCGCTGCTCAAGTTCTTGGTCACCAATGAAACCGCGTTACTGAAGCGACACAATATCTTCATAATGGCGCAACAGGTGGTGAACATTTTCAACTTATTTATAACATTGGGCGACACGTTCCTACCCACTCCGGCATCGTACGATCAATTATACTACGAGCTAATTAGAATGCATCAAGTTTTCGATAATCTATTTTATATGGCTCTGCGGTACTCCACCGGCGACGGCGAATATAAAGCCGAAGCGTTGCGACTGGCCAATTGCTTAGTGAACGTGCGCGCCATCATCCAGCACTTCTCCGTGAAGATAGAAGCGTGGCTCGACCAACAGAATCTGTCGACTCCCTCCGAGGAGCAGATACTGGAAGTGGTACGCAAGAACTACGACAGCCTGATACTGAAGCTACAGAAGGGACTCGAGAGCTATGAACGCTATTCGGAGCAAGAGCACTGGCCGCTGCTGGCGAGGACGGTGCGGGCGGCGCGGAGGAGGGGGGACGCCAGCGCGCTGCAGCAGCACTCCGCCGCCGCGCTCCACCACTACACTGCCGCCCCGTGA
Protein
MAMRKRSGSGSKRHLKEKVVQIYESFFRGEDLTTDNPTFWDEFFLLKPKIPQLEAEIHKLSTEQLINMKESMNILVEQCIEMLRQDHHIRLLYALQTLSALLITMYQRLEQDSNINIKTIMIGQENADTKMVKLFEQCSIILSGDVHESLKSMVLKLLLIIATGIENIDENPFVEYFMSNCLFEPLIQLLCTTTERQQHGYDVVMLLMFLINYKKHESTNPYVVKLSILDDELALNGYGQVITTALNDYVMSAFGGMQGNGGGGGWLSSLTSMVGGIFLTNDEQQPLARGQRNNGAEEGMLLAMYEASHLNRNFMTTLAHSSSTTASSAPPSPPATLPPHQSPPNLAQIQALDNDQPTNLLVTFFQYCSIIMADTRSESSINKCSLCFITLTCIVEDQFANSIMHDQNLTFKVQLYRLPMRHRKIVLEEPPSQPLASTLIDLLVEFIMCHLLKKFPGELYSLCVGVLLRLLSYQKRCKVRLARDWRPLWAALIALLKFLVTNETALLKRHNIFIMAQQVVNIFNLFITLGDTFLPTPASYDQLYYELIRMHQVFDNLFYMALRYSTGDGEYKAEALRLANCLVNVRAIIQHFSVKIEAWLDQQNLSTPSEEQILEVVRKNYDSLILKLQKGLESYERYSEQEHWPLLARTVRAARRRGDASALQQHSAAALHHYTAAP
Summary
Uniprot
A0A2A4JQQ1
A0A2H1VNB8
A0A1E1WKT1
A0A0N1I5A2
A0A194RPR7
A0A067QN84
+ More
A0A2J7PBZ8 A0A1B6FLB7 A0A1Y1NCJ2 E0V948 A0A023F2T1 A0A154PKQ7 A0A088ADG0 E2BL44 A0A026VW15 A0A195C8A2 A0A146L9F2 F4WLY4 A0A195FU59 A0A195BCJ2 A0A158N9T3 A0A195DCV1 U4U2H9 E9HHW9 A0A0P5B652 A0A0P5B650 A0A0P4YHH9 A0A0P5V5M0 A0A0P5XJH7 A0A0P5U1Z7 A0A0P5YEV4 A0A0P5VG04 A0A0P4WJU4 A0A0P5PNG1 A0A1B0C5J4 A0A0V0GAT8 A0A1A9XZV5 D6X3W0 A0A0P5X091 A0A0P6B0U4 A0A1B0FMF2 A0A1L8DCN4 A0A1A9Z6F6 A0A1A9VA25 A0A1S3HJ05 A0A0P4W2A3 A0A2J7PC06 B4N813 A0A0R1E5T9 Q9VHK5 R7UND4 A0A1W4W2Q9 B4LXK3 B4K9D8 A0A3B0JTP1 B3P1W8 B4PTX4 Q295Q1 Q7KSU3 A0A1W4VQ07 T1HGG5 B4HKZ3 B4GFV6 A0A0K8W260 W8BKJ6 B4JTI6 A0A0K8TR82 A0A0A1XKS8 A0A1I8P687 T1PHT6 A0A034WRT2 A0A210PPF7 A0A1I8NES1 A0A0M3QXH1 A7RII7 A0A1E1X9B5 A0A0B6ZUJ8 A0A0P5XB07 B4QXW6 B3LZ02 A0A224Y488 A0A131YJL9 A0A131X9Z7 A0A0K8RGV9 L7M1M2 A0A1E1XU46 A0A023FNS8 A0A0L8GJP6 A0A1S4FUC6 Q16NY2 G3MQ91 A0A224XLS7 A0A1B6C119 A0A023EVS2 A0A3B3SZW3 L7LTV9 A0A0L7RJ71 B7PL50 A0A1L8FJD3
A0A2J7PBZ8 A0A1B6FLB7 A0A1Y1NCJ2 E0V948 A0A023F2T1 A0A154PKQ7 A0A088ADG0 E2BL44 A0A026VW15 A0A195C8A2 A0A146L9F2 F4WLY4 A0A195FU59 A0A195BCJ2 A0A158N9T3 A0A195DCV1 U4U2H9 E9HHW9 A0A0P5B652 A0A0P5B650 A0A0P4YHH9 A0A0P5V5M0 A0A0P5XJH7 A0A0P5U1Z7 A0A0P5YEV4 A0A0P5VG04 A0A0P4WJU4 A0A0P5PNG1 A0A1B0C5J4 A0A0V0GAT8 A0A1A9XZV5 D6X3W0 A0A0P5X091 A0A0P6B0U4 A0A1B0FMF2 A0A1L8DCN4 A0A1A9Z6F6 A0A1A9VA25 A0A1S3HJ05 A0A0P4W2A3 A0A2J7PC06 B4N813 A0A0R1E5T9 Q9VHK5 R7UND4 A0A1W4W2Q9 B4LXK3 B4K9D8 A0A3B0JTP1 B3P1W8 B4PTX4 Q295Q1 Q7KSU3 A0A1W4VQ07 T1HGG5 B4HKZ3 B4GFV6 A0A0K8W260 W8BKJ6 B4JTI6 A0A0K8TR82 A0A0A1XKS8 A0A1I8P687 T1PHT6 A0A034WRT2 A0A210PPF7 A0A1I8NES1 A0A0M3QXH1 A7RII7 A0A1E1X9B5 A0A0B6ZUJ8 A0A0P5XB07 B4QXW6 B3LZ02 A0A224Y488 A0A131YJL9 A0A131X9Z7 A0A0K8RGV9 L7M1M2 A0A1E1XU46 A0A023FNS8 A0A0L8GJP6 A0A1S4FUC6 Q16NY2 G3MQ91 A0A224XLS7 A0A1B6C119 A0A023EVS2 A0A3B3SZW3 L7LTV9 A0A0L7RJ71 B7PL50 A0A1L8FJD3
Pubmed
26354079
24845553
28004739
20566863
25474469
20798317
+ More
24508170 30249741 26823975 21719571 21347285 23537049 21292972 18362917 19820115 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23254933 15632085 24495485 26369729 25830018 25348373 28812685 25315136 17615350 28503490 28797301 26830274 28049606 25576852 29209593 17510324 22216098 24945155 29240929 27762356
24508170 30249741 26823975 21719571 21347285 23537049 21292972 18362917 19820115 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23254933 15632085 24495485 26369729 25830018 25348373 28812685 25315136 17615350 28503490 28797301 26830274 28049606 25576852 29209593 17510324 22216098 24945155 29240929 27762356
EMBL
NWSH01000798
PCG74139.1
ODYU01003335
SOQ41952.1
GDQN01003431
JAT87623.1
+ More
KQ459037 KPJ04282.1 KQ459833 KPJ19848.1 KK853137 KDR10796.1 NEVH01027073 PNF13863.1 GECZ01018808 JAS50961.1 GEZM01006882 JAV95603.1 DS234988 EEB09904.1 GBBI01003373 JAC15339.1 KQ434948 KZC12436.1 GL448921 EFN83594.1 KK107796 QOIP01000013 EZA47686.1 RLU15185.1 KQ978220 KYM96363.1 GDHC01014773 GDHC01010398 JAQ03856.1 JAQ08231.1 GL888217 EGI64628.1 KQ981264 KYN43961.1 KQ976528 KYM81914.1 ADTU01001412 KQ980989 KYN10662.1 KB631815 ERL86513.1 GL732651 EFX68666.1 GDIP01189026 JAJ34376.1 GDIP01189027 JAJ34375.1 GDIP01227622 JAI95779.1 GDIP01141596 GDIP01105035 LRGB01000491 JAL98679.1 KZS18963.1 GDIP01071144 JAM32571.1 GDIP01121131 JAL82583.1 GDIP01058849 JAM44866.1 GDIP01102190 JAM01525.1 GDIP01254365 GDIQ01052560 JAI69036.1 JAN42177.1 GDIQ01125925 JAL25801.1 JXJN01026043 GECL01000955 JAP05169.1 KQ971379 EEZ97362.1 GDIP01079581 JAM24134.1 GDIP01022662 JAM81053.1 CCAG010016541 GFDF01009868 JAV04216.1 GDRN01094585 GDRN01094584 JAI59702.1 PNF13866.1 CH964232 EDW81264.1 CM000160 KRK03201.1 AE014297 AY069483 AAF54299.2 AAL39628.1 AMQN01007821 KB301483 ELU05457.1 CH940650 EDW66787.1 CH933806 EDW15570.1 OUUW01000008 SPP84423.1 CH954181 EDV49856.1 EDW96585.1 CM000070 EAL28657.2 BT128739 AAS65128.1 AEH59642.1 ACPB03015959 CH480815 EDW42957.1 CH479182 EDW34491.1 GDHF01007130 JAI45184.1 GAMC01016486 JAB90069.1 CH916373 EDV95076.1 GDAI01000967 JAI16636.1 GBXI01002686 JAD11606.1 KA648377 AFP63006.1 GAKP01001608 JAC57344.1 NEDP02005569 OWF38357.1 CP012526 ALC45859.1 DS469512 EDO48716.1 GFAC01003360 JAT95828.1 HACG01025449 CEK72314.1 GDIP01075782 JAM27933.1 CM000364 EDX13703.1 CH902617 EDV41876.1 GFPF01001231 MAA12377.1 GEDV01009064 JAP79493.1 GEFH01004932 JAP63649.1 GADI01003433 JAA70375.1 GACK01006828 JAA58206.1 GFAA01000606 JAU02829.1 GBBK01002159 JAC22323.1 KQ421523 KOF77226.1 CH477803 EAT36060.1 JO844042 AEO35659.1 GFTR01007323 JAW09103.1 GEDC01030234 JAS07064.1 GAPW01000422 JAC13176.1 GACK01010002 JAA55032.1 KQ414581 KOC71017.1 ABJB010022857 ABJB010087777 ABJB010112357 ABJB010217244 ABJB010218823 ABJB010258738 ABJB010472833 ABJB010578851 ABJB010685015 ABJB011006260 DS737809 EEC07322.1 CM004478 OCT71693.1
KQ459037 KPJ04282.1 KQ459833 KPJ19848.1 KK853137 KDR10796.1 NEVH01027073 PNF13863.1 GECZ01018808 JAS50961.1 GEZM01006882 JAV95603.1 DS234988 EEB09904.1 GBBI01003373 JAC15339.1 KQ434948 KZC12436.1 GL448921 EFN83594.1 KK107796 QOIP01000013 EZA47686.1 RLU15185.1 KQ978220 KYM96363.1 GDHC01014773 GDHC01010398 JAQ03856.1 JAQ08231.1 GL888217 EGI64628.1 KQ981264 KYN43961.1 KQ976528 KYM81914.1 ADTU01001412 KQ980989 KYN10662.1 KB631815 ERL86513.1 GL732651 EFX68666.1 GDIP01189026 JAJ34376.1 GDIP01189027 JAJ34375.1 GDIP01227622 JAI95779.1 GDIP01141596 GDIP01105035 LRGB01000491 JAL98679.1 KZS18963.1 GDIP01071144 JAM32571.1 GDIP01121131 JAL82583.1 GDIP01058849 JAM44866.1 GDIP01102190 JAM01525.1 GDIP01254365 GDIQ01052560 JAI69036.1 JAN42177.1 GDIQ01125925 JAL25801.1 JXJN01026043 GECL01000955 JAP05169.1 KQ971379 EEZ97362.1 GDIP01079581 JAM24134.1 GDIP01022662 JAM81053.1 CCAG010016541 GFDF01009868 JAV04216.1 GDRN01094585 GDRN01094584 JAI59702.1 PNF13866.1 CH964232 EDW81264.1 CM000160 KRK03201.1 AE014297 AY069483 AAF54299.2 AAL39628.1 AMQN01007821 KB301483 ELU05457.1 CH940650 EDW66787.1 CH933806 EDW15570.1 OUUW01000008 SPP84423.1 CH954181 EDV49856.1 EDW96585.1 CM000070 EAL28657.2 BT128739 AAS65128.1 AEH59642.1 ACPB03015959 CH480815 EDW42957.1 CH479182 EDW34491.1 GDHF01007130 JAI45184.1 GAMC01016486 JAB90069.1 CH916373 EDV95076.1 GDAI01000967 JAI16636.1 GBXI01002686 JAD11606.1 KA648377 AFP63006.1 GAKP01001608 JAC57344.1 NEDP02005569 OWF38357.1 CP012526 ALC45859.1 DS469512 EDO48716.1 GFAC01003360 JAT95828.1 HACG01025449 CEK72314.1 GDIP01075782 JAM27933.1 CM000364 EDX13703.1 CH902617 EDV41876.1 GFPF01001231 MAA12377.1 GEDV01009064 JAP79493.1 GEFH01004932 JAP63649.1 GADI01003433 JAA70375.1 GACK01006828 JAA58206.1 GFAA01000606 JAU02829.1 GBBK01002159 JAC22323.1 KQ421523 KOF77226.1 CH477803 EAT36060.1 JO844042 AEO35659.1 GFTR01007323 JAW09103.1 GEDC01030234 JAS07064.1 GAPW01000422 JAC13176.1 GACK01010002 JAA55032.1 KQ414581 KOC71017.1 ABJB010022857 ABJB010087777 ABJB010112357 ABJB010217244 ABJB010218823 ABJB010258738 ABJB010472833 ABJB010578851 ABJB010685015 ABJB011006260 DS737809 EEC07322.1 CM004478 OCT71693.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000027135
UP000235965
UP000009046
+ More
UP000076502 UP000005203 UP000008237 UP000053097 UP000279307 UP000078542 UP000007755 UP000078541 UP000078540 UP000005205 UP000078492 UP000030742 UP000000305 UP000076858 UP000092460 UP000092443 UP000007266 UP000092444 UP000092445 UP000078200 UP000085678 UP000007798 UP000002282 UP000000803 UP000014760 UP000192221 UP000008792 UP000009192 UP000268350 UP000008711 UP000001819 UP000015103 UP000001292 UP000008744 UP000001070 UP000095300 UP000242188 UP000095301 UP000092553 UP000001593 UP000000304 UP000007801 UP000053454 UP000008820 UP000261540 UP000053825 UP000001555 UP000186698
UP000076502 UP000005203 UP000008237 UP000053097 UP000279307 UP000078542 UP000007755 UP000078541 UP000078540 UP000005205 UP000078492 UP000030742 UP000000305 UP000076858 UP000092460 UP000092443 UP000007266 UP000092444 UP000092445 UP000078200 UP000085678 UP000007798 UP000002282 UP000000803 UP000014760 UP000192221 UP000008792 UP000009192 UP000268350 UP000008711 UP000001819 UP000015103 UP000001292 UP000008744 UP000001070 UP000095300 UP000242188 UP000095301 UP000092553 UP000001593 UP000000304 UP000007801 UP000053454 UP000008820 UP000261540 UP000053825 UP000001555 UP000186698
Pfam
PF08427 DUF1741
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A2A4JQQ1
A0A2H1VNB8
A0A1E1WKT1
A0A0N1I5A2
A0A194RPR7
A0A067QN84
+ More
A0A2J7PBZ8 A0A1B6FLB7 A0A1Y1NCJ2 E0V948 A0A023F2T1 A0A154PKQ7 A0A088ADG0 E2BL44 A0A026VW15 A0A195C8A2 A0A146L9F2 F4WLY4 A0A195FU59 A0A195BCJ2 A0A158N9T3 A0A195DCV1 U4U2H9 E9HHW9 A0A0P5B652 A0A0P5B650 A0A0P4YHH9 A0A0P5V5M0 A0A0P5XJH7 A0A0P5U1Z7 A0A0P5YEV4 A0A0P5VG04 A0A0P4WJU4 A0A0P5PNG1 A0A1B0C5J4 A0A0V0GAT8 A0A1A9XZV5 D6X3W0 A0A0P5X091 A0A0P6B0U4 A0A1B0FMF2 A0A1L8DCN4 A0A1A9Z6F6 A0A1A9VA25 A0A1S3HJ05 A0A0P4W2A3 A0A2J7PC06 B4N813 A0A0R1E5T9 Q9VHK5 R7UND4 A0A1W4W2Q9 B4LXK3 B4K9D8 A0A3B0JTP1 B3P1W8 B4PTX4 Q295Q1 Q7KSU3 A0A1W4VQ07 T1HGG5 B4HKZ3 B4GFV6 A0A0K8W260 W8BKJ6 B4JTI6 A0A0K8TR82 A0A0A1XKS8 A0A1I8P687 T1PHT6 A0A034WRT2 A0A210PPF7 A0A1I8NES1 A0A0M3QXH1 A7RII7 A0A1E1X9B5 A0A0B6ZUJ8 A0A0P5XB07 B4QXW6 B3LZ02 A0A224Y488 A0A131YJL9 A0A131X9Z7 A0A0K8RGV9 L7M1M2 A0A1E1XU46 A0A023FNS8 A0A0L8GJP6 A0A1S4FUC6 Q16NY2 G3MQ91 A0A224XLS7 A0A1B6C119 A0A023EVS2 A0A3B3SZW3 L7LTV9 A0A0L7RJ71 B7PL50 A0A1L8FJD3
A0A2J7PBZ8 A0A1B6FLB7 A0A1Y1NCJ2 E0V948 A0A023F2T1 A0A154PKQ7 A0A088ADG0 E2BL44 A0A026VW15 A0A195C8A2 A0A146L9F2 F4WLY4 A0A195FU59 A0A195BCJ2 A0A158N9T3 A0A195DCV1 U4U2H9 E9HHW9 A0A0P5B652 A0A0P5B650 A0A0P4YHH9 A0A0P5V5M0 A0A0P5XJH7 A0A0P5U1Z7 A0A0P5YEV4 A0A0P5VG04 A0A0P4WJU4 A0A0P5PNG1 A0A1B0C5J4 A0A0V0GAT8 A0A1A9XZV5 D6X3W0 A0A0P5X091 A0A0P6B0U4 A0A1B0FMF2 A0A1L8DCN4 A0A1A9Z6F6 A0A1A9VA25 A0A1S3HJ05 A0A0P4W2A3 A0A2J7PC06 B4N813 A0A0R1E5T9 Q9VHK5 R7UND4 A0A1W4W2Q9 B4LXK3 B4K9D8 A0A3B0JTP1 B3P1W8 B4PTX4 Q295Q1 Q7KSU3 A0A1W4VQ07 T1HGG5 B4HKZ3 B4GFV6 A0A0K8W260 W8BKJ6 B4JTI6 A0A0K8TR82 A0A0A1XKS8 A0A1I8P687 T1PHT6 A0A034WRT2 A0A210PPF7 A0A1I8NES1 A0A0M3QXH1 A7RII7 A0A1E1X9B5 A0A0B6ZUJ8 A0A0P5XB07 B4QXW6 B3LZ02 A0A224Y488 A0A131YJL9 A0A131X9Z7 A0A0K8RGV9 L7M1M2 A0A1E1XU46 A0A023FNS8 A0A0L8GJP6 A0A1S4FUC6 Q16NY2 G3MQ91 A0A224XLS7 A0A1B6C119 A0A023EVS2 A0A3B3SZW3 L7LTV9 A0A0L7RJ71 B7PL50 A0A1L8FJD3
Ontologies
GO
PANTHER
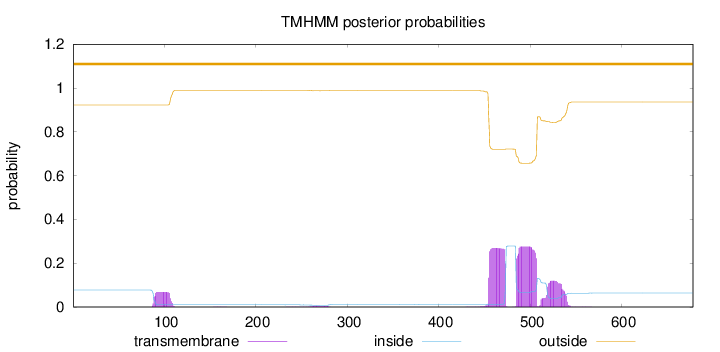
Topology
Length:
678
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.96595
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07738
outside
1 - 678
Population Genetic Test Statistics
Pi
243.140856
Theta
185.321157
Tajima's D
1.098472
CLR
0.352632
CSRT
0.689465526723664
Interpretation
Uncertain
