Pre Gene Modal
BGIBMGA006641
Annotation
ras-related_GTP-binding_protein_4b_[Bombyx_mori]
Full name
Dihydroorotate dehydrogenase (quinone), mitochondrial
Location in the cell
Mitochondrial Reliability : 1.295
Sequence
CDS
ATGTCTGAATCGTACGAATATCTTTTCAAATTTCTTGTGATTGGAAGTGCAGGCACAGGAAAGTCAAGCTTACTAAATAATTTTATCGGAAATAAATTCAAGGAAGATCGATGTCACACTATTGGTGTTGAATTTGGTTCCAAAATTGTGAACATCGGTGGCAAATCAACTAAGTTGCAAATATGGGACACAGCTGGACAGGAACGGTTTCGATCTGTGACCAGATCATATTACCGCGGCGCAGCAGGAGCTCTGCTAGTGTATGATATTACATCTAGAGATTCATTCAATGCTCTTGCTAATTGGCTGAGAGATGCTCGCACTCTTGCAAGTCCTAATATCGTAATTTTGCTTGTGGGCAACAAGAAAGACTTAGAAGATTCCAGAGAAGTAACTTTTACTGAGGCAAGTCAGTTTGCTCTTGAAAATGAATTAATGTTTTTAGAAACAAGTGCTAAAACAGGAGAAAATGTTGAAGAAGCTTTTCTGAAATGCTCGAAAACAATTCTAGCTAAGATTGAAACTGGTGAATTGGATCCTGAAAGGATAGGTTCTGGTATTCAATATGGCACAAATACTACAAAGAGATTGACTGCTCCAAAGAGGCCAGCTAGAACCCCTTCAGAATGTGCTTGCCGTGTTTAA
Protein
MSESYEYLFKFLVIGSAGTGKSSLLNNFIGNKFKEDRCHTIGVEFGSKIVNIGGKSTKLQIWDTAGQERFRSVTRSYYRGAAGALLVYDITSRDSFNALANWLRDARTLASPNIVILLVGNKKDLEDSREVTFTEASQFALENELMFLETSAKTGENVEEAFLKCSKTILAKIETGELDPERIGSGIQYGTNTTKRLTAPKRPARTPSECACRV
Summary
Catalytic Activity
(S)-dihydroorotate + a quinone = a quinol + orotate
Cofactor
FMN
Similarity
Belongs to the dihydroorotate dehydrogenase family. Type 2 subfamily.
Uniprot
Q2F5W7
A0A2A4JRQ9
I3PL64
A0A2W1BEG0
A0A194RPR2
A0A212FE69
+ More
A0A0K8TNV1 V5HWI8 A0A0A1WR81 A0A0K8UXV9 A0A034W9F3 A0A1I8Q9L5 G3MNH8 W8BND9 B4MIR7 A0A0N0P9Y4 A0A1B0GIN8 A0A1L8DV24 A0A1B0DL31 A0A0L0CH89 A0A224Z5J1 A0A131YU67 L7M4Y9 A0A131XET0 T1PG75 A0A067R2L0 A0A0M4EBT2 B4J4B2 A0A1W4VD77 A0A3B0JW07 Q291A4 B4GAZ9 B4P5E4 B3MIH3 B4HMX3 B3NMJ8 B4QBA1 Q7KY04 A0A2H1VTV9 B4KM01 B5RJE3 B4LIJ3 D3TP13 A0A1A9XWU7 A0A1A9VGP9 A0A1B0BAZ9 A0A1A9ZWT9 A0A087TB09 B7Q2W4 A0A2P8XX71 A0A0L7LJ17 A0A2R5LNQ3 A0A1Z5KYH7 A0A293LIE1 A0A1E1X5X4 A0A023FFP5 A0A1B6BX20 R7VCQ9 A0A0P4WC93 Q16SZ9 H9GK45 A0A3B4BXK3 W5NCH5 W5LM81 A0A182JLU4 A0A2J7PNS5 U5EJ77 A0A182Y5G8 A0A182UWJ2 A0A182U4A5 A0A182L773 A0A182XG84 Q7QB34 A0A182IDC9 A0A1Q3FDX9 G1K2H7 A0A151PE10 A0A182R0M6 W4YGM6 A0A182LV58 A0A1W5A2T8 A0A210PPX6 A0A182RZJ2 B0WU66 A0A182F7T8 G3TWS7 A0A2Y9ECN0 A0A023EMC6 A0A182NLF8 W5JLQ1 E3TEK2 A0A1B6MQC3 A0A2M4ACH7 A0A2M4BZ25 T1E7B2 A0A2M3Z7X2 H2U0D4 A0A182PLV9 G1U1H3 H3CQN7
A0A0K8TNV1 V5HWI8 A0A0A1WR81 A0A0K8UXV9 A0A034W9F3 A0A1I8Q9L5 G3MNH8 W8BND9 B4MIR7 A0A0N0P9Y4 A0A1B0GIN8 A0A1L8DV24 A0A1B0DL31 A0A0L0CH89 A0A224Z5J1 A0A131YU67 L7M4Y9 A0A131XET0 T1PG75 A0A067R2L0 A0A0M4EBT2 B4J4B2 A0A1W4VD77 A0A3B0JW07 Q291A4 B4GAZ9 B4P5E4 B3MIH3 B4HMX3 B3NMJ8 B4QBA1 Q7KY04 A0A2H1VTV9 B4KM01 B5RJE3 B4LIJ3 D3TP13 A0A1A9XWU7 A0A1A9VGP9 A0A1B0BAZ9 A0A1A9ZWT9 A0A087TB09 B7Q2W4 A0A2P8XX71 A0A0L7LJ17 A0A2R5LNQ3 A0A1Z5KYH7 A0A293LIE1 A0A1E1X5X4 A0A023FFP5 A0A1B6BX20 R7VCQ9 A0A0P4WC93 Q16SZ9 H9GK45 A0A3B4BXK3 W5NCH5 W5LM81 A0A182JLU4 A0A2J7PNS5 U5EJ77 A0A182Y5G8 A0A182UWJ2 A0A182U4A5 A0A182L773 A0A182XG84 Q7QB34 A0A182IDC9 A0A1Q3FDX9 G1K2H7 A0A151PE10 A0A182R0M6 W4YGM6 A0A182LV58 A0A1W5A2T8 A0A210PPX6 A0A182RZJ2 B0WU66 A0A182F7T8 G3TWS7 A0A2Y9ECN0 A0A023EMC6 A0A182NLF8 W5JLQ1 E3TEK2 A0A1B6MQC3 A0A2M4ACH7 A0A2M4BZ25 T1E7B2 A0A2M3Z7X2 H2U0D4 A0A182PLV9 G1U1H3 H3CQN7
EC Number
1.3.5.2
Pubmed
19121390
28756777
26354079
22118469
26369729
25765539
+ More
25830018 25348373 22216098 24495485 17994087 26108605 28797301 26830274 25576852 28049606 25315136 24845553 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 29403074 26227816 28528879 28503490 23254933 17510324 21881562 25329095 25244985 20966253 12364791 14747013 17210077 23594743 22293439 28812685 24945155 20920257 23761445 20634964 21551351 21993624 15496914
25830018 25348373 22216098 24495485 17994087 26108605 28797301 26830274 25576852 28049606 25315136 24845553 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 29403074 26227816 28528879 28503490 23254933 17510324 21881562 25329095 25244985 20966253 12364791 14747013 17210077 23594743 22293439 28812685 24945155 20920257 23761445 20634964 21551351 21993624 15496914
EMBL
BABH01008418
DQ311306
ABD36250.1
NWSH01000798
PCG74153.1
JN634855
+ More
AFA36651.1 KZ150160 PZC72721.1 KQ459833 KPJ19843.1 AGBW02008970 OWR52032.1 GDAI01001559 JAI16044.1 GANP01005645 JAB78823.1 GBXI01013141 JAD01151.1 GDHF01020951 GDHF01003666 JAI31363.1 JAI48648.1 GAKP01008001 GAKP01008000 JAC50952.1 JO843429 AEO35046.1 GAMC01006218 JAC00338.1 CH963719 EDW72006.1 KQ459037 KPJ04276.1 AJWK01015122 AJWK01015123 GFDF01003797 JAV10287.1 AJVK01069137 JRES01000408 KNC31615.1 GFPF01011215 MAA22361.1 GEDV01005784 JAP82773.1 GACK01005959 JAA59075.1 GEFH01003991 JAP64590.1 KA647684 AFP62313.1 KK852747 KDR17258.1 CP012524 ALC41052.1 CH916367 EDW00592.1 OUUW01000001 SPP75258.1 CM000071 EAL25208.2 CH479181 EDW32101.1 CM000158 EDW91775.1 CH902619 EDV37021.1 CH480816 EDW48323.1 CH954179 EDV54937.1 CM000362 CM002911 EDX07525.1 KMY94628.1 AE013599 AB035670 AAF57831.1 AAM68480.1 BAA88243.1 ODYU01004426 SOQ44281.1 CH933808 EDW10790.1 BT044417 ACH92482.1 CH940648 EDW60363.1 EZ423165 ADD19441.1 JXJN01011233 KK114378 KFM62298.1 ABJB010347534 ABJB010524691 ABJB010757334 DS846142 EEC13186.1 PYGN01001224 PSN36514.1 JTDY01000888 KOB75553.1 GGLE01007040 MBY11166.1 GFJQ02006793 JAW00177.1 GFWV01003544 GFWV01023018 MAA28274.1 GFAC01004519 JAT94669.1 GBBK01003681 JAC20801.1 GEDC01031748 JAS05550.1 AMQN01004321 KB293181 ELU16342.1 GDRN01074760 GDRN01074759 JAI63177.1 CH477662 EAT37591.1 AHAT01023914 AHAT01023915 NEVH01023316 PNF17978.1 GANO01002339 JAB57532.1 AAAB01008880 EAA08616.2 APCN01005106 GFDL01009316 JAV25729.1 CR384088 AKHW03000482 KYO47164.1 AXCN02000134 AAGJ04037203 AXCM01006741 NEDP02005562 OWF38555.1 DS232101 EDS34786.1 GAPW01004134 JAC09464.1 ADMH02000787 ETN65056.1 GU588784 ADO28738.1 GEBQ01001878 JAT38099.1 GGFK01005162 MBW38483.1 GGFJ01009113 MBW58254.1 GAMD01003192 JAA98398.1 GGFM01003866 MBW24617.1
AFA36651.1 KZ150160 PZC72721.1 KQ459833 KPJ19843.1 AGBW02008970 OWR52032.1 GDAI01001559 JAI16044.1 GANP01005645 JAB78823.1 GBXI01013141 JAD01151.1 GDHF01020951 GDHF01003666 JAI31363.1 JAI48648.1 GAKP01008001 GAKP01008000 JAC50952.1 JO843429 AEO35046.1 GAMC01006218 JAC00338.1 CH963719 EDW72006.1 KQ459037 KPJ04276.1 AJWK01015122 AJWK01015123 GFDF01003797 JAV10287.1 AJVK01069137 JRES01000408 KNC31615.1 GFPF01011215 MAA22361.1 GEDV01005784 JAP82773.1 GACK01005959 JAA59075.1 GEFH01003991 JAP64590.1 KA647684 AFP62313.1 KK852747 KDR17258.1 CP012524 ALC41052.1 CH916367 EDW00592.1 OUUW01000001 SPP75258.1 CM000071 EAL25208.2 CH479181 EDW32101.1 CM000158 EDW91775.1 CH902619 EDV37021.1 CH480816 EDW48323.1 CH954179 EDV54937.1 CM000362 CM002911 EDX07525.1 KMY94628.1 AE013599 AB035670 AAF57831.1 AAM68480.1 BAA88243.1 ODYU01004426 SOQ44281.1 CH933808 EDW10790.1 BT044417 ACH92482.1 CH940648 EDW60363.1 EZ423165 ADD19441.1 JXJN01011233 KK114378 KFM62298.1 ABJB010347534 ABJB010524691 ABJB010757334 DS846142 EEC13186.1 PYGN01001224 PSN36514.1 JTDY01000888 KOB75553.1 GGLE01007040 MBY11166.1 GFJQ02006793 JAW00177.1 GFWV01003544 GFWV01023018 MAA28274.1 GFAC01004519 JAT94669.1 GBBK01003681 JAC20801.1 GEDC01031748 JAS05550.1 AMQN01004321 KB293181 ELU16342.1 GDRN01074760 GDRN01074759 JAI63177.1 CH477662 EAT37591.1 AHAT01023914 AHAT01023915 NEVH01023316 PNF17978.1 GANO01002339 JAB57532.1 AAAB01008880 EAA08616.2 APCN01005106 GFDL01009316 JAV25729.1 CR384088 AKHW03000482 KYO47164.1 AXCN02000134 AAGJ04037203 AXCM01006741 NEDP02005562 OWF38555.1 DS232101 EDS34786.1 GAPW01004134 JAC09464.1 ADMH02000787 ETN65056.1 GU588784 ADO28738.1 GEBQ01001878 JAT38099.1 GGFK01005162 MBW38483.1 GGFJ01009113 MBW58254.1 GAMD01003192 JAA98398.1 GGFM01003866 MBW24617.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000095300
UP000007798
+ More
UP000053268 UP000092461 UP000092462 UP000037069 UP000095301 UP000027135 UP000092553 UP000001070 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000008792 UP000092443 UP000078200 UP000092460 UP000092445 UP000054359 UP000001555 UP000245037 UP000037510 UP000014760 UP000008820 UP000001646 UP000261440 UP000018468 UP000018467 UP000075880 UP000235965 UP000076408 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000437 UP000050525 UP000075886 UP000007110 UP000075883 UP000192224 UP000242188 UP000075900 UP000002320 UP000069272 UP000007646 UP000248480 UP000075884 UP000000673 UP000221080 UP000005226 UP000075885 UP000001811 UP000007303
UP000053268 UP000092461 UP000092462 UP000037069 UP000095301 UP000027135 UP000092553 UP000001070 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000009192 UP000008792 UP000092443 UP000078200 UP000092460 UP000092445 UP000054359 UP000001555 UP000245037 UP000037510 UP000014760 UP000008820 UP000001646 UP000261440 UP000018468 UP000018467 UP000075880 UP000235965 UP000076408 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000437 UP000050525 UP000075886 UP000007110 UP000075883 UP000192224 UP000242188 UP000075900 UP000002320 UP000069272 UP000007646 UP000248480 UP000075884 UP000000673 UP000221080 UP000005226 UP000075885 UP000001811 UP000007303
Interpro
IPR027417
P-loop_NTPase
+ More
IPR001806 Small_GTPase
IPR041819 Rab4
IPR005225 Small_GTP-bd_dom
IPR013785 Aldolase_TIM
IPR005719 Dihydroorotate_DH_2
IPR001295 Dihydroorotate_DH_CS
IPR005720 Dihydroorotate_DH
IPR036554 GHMP_kinase_C_sf
IPR029765 Mev_diP_decarb
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR020568 Ribosomal_S5_D2-typ_fold
IPR041431 Mvd1_C
IPR001806 Small_GTPase
IPR041819 Rab4
IPR005225 Small_GTP-bd_dom
IPR013785 Aldolase_TIM
IPR005719 Dihydroorotate_DH_2
IPR001295 Dihydroorotate_DH_CS
IPR005720 Dihydroorotate_DH
IPR036554 GHMP_kinase_C_sf
IPR029765 Mev_diP_decarb
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR020568 Ribosomal_S5_D2-typ_fold
IPR041431 Mvd1_C
Gene 3D
ProteinModelPortal
Q2F5W7
A0A2A4JRQ9
I3PL64
A0A2W1BEG0
A0A194RPR2
A0A212FE69
+ More
A0A0K8TNV1 V5HWI8 A0A0A1WR81 A0A0K8UXV9 A0A034W9F3 A0A1I8Q9L5 G3MNH8 W8BND9 B4MIR7 A0A0N0P9Y4 A0A1B0GIN8 A0A1L8DV24 A0A1B0DL31 A0A0L0CH89 A0A224Z5J1 A0A131YU67 L7M4Y9 A0A131XET0 T1PG75 A0A067R2L0 A0A0M4EBT2 B4J4B2 A0A1W4VD77 A0A3B0JW07 Q291A4 B4GAZ9 B4P5E4 B3MIH3 B4HMX3 B3NMJ8 B4QBA1 Q7KY04 A0A2H1VTV9 B4KM01 B5RJE3 B4LIJ3 D3TP13 A0A1A9XWU7 A0A1A9VGP9 A0A1B0BAZ9 A0A1A9ZWT9 A0A087TB09 B7Q2W4 A0A2P8XX71 A0A0L7LJ17 A0A2R5LNQ3 A0A1Z5KYH7 A0A293LIE1 A0A1E1X5X4 A0A023FFP5 A0A1B6BX20 R7VCQ9 A0A0P4WC93 Q16SZ9 H9GK45 A0A3B4BXK3 W5NCH5 W5LM81 A0A182JLU4 A0A2J7PNS5 U5EJ77 A0A182Y5G8 A0A182UWJ2 A0A182U4A5 A0A182L773 A0A182XG84 Q7QB34 A0A182IDC9 A0A1Q3FDX9 G1K2H7 A0A151PE10 A0A182R0M6 W4YGM6 A0A182LV58 A0A1W5A2T8 A0A210PPX6 A0A182RZJ2 B0WU66 A0A182F7T8 G3TWS7 A0A2Y9ECN0 A0A023EMC6 A0A182NLF8 W5JLQ1 E3TEK2 A0A1B6MQC3 A0A2M4ACH7 A0A2M4BZ25 T1E7B2 A0A2M3Z7X2 H2U0D4 A0A182PLV9 G1U1H3 H3CQN7
A0A0K8TNV1 V5HWI8 A0A0A1WR81 A0A0K8UXV9 A0A034W9F3 A0A1I8Q9L5 G3MNH8 W8BND9 B4MIR7 A0A0N0P9Y4 A0A1B0GIN8 A0A1L8DV24 A0A1B0DL31 A0A0L0CH89 A0A224Z5J1 A0A131YU67 L7M4Y9 A0A131XET0 T1PG75 A0A067R2L0 A0A0M4EBT2 B4J4B2 A0A1W4VD77 A0A3B0JW07 Q291A4 B4GAZ9 B4P5E4 B3MIH3 B4HMX3 B3NMJ8 B4QBA1 Q7KY04 A0A2H1VTV9 B4KM01 B5RJE3 B4LIJ3 D3TP13 A0A1A9XWU7 A0A1A9VGP9 A0A1B0BAZ9 A0A1A9ZWT9 A0A087TB09 B7Q2W4 A0A2P8XX71 A0A0L7LJ17 A0A2R5LNQ3 A0A1Z5KYH7 A0A293LIE1 A0A1E1X5X4 A0A023FFP5 A0A1B6BX20 R7VCQ9 A0A0P4WC93 Q16SZ9 H9GK45 A0A3B4BXK3 W5NCH5 W5LM81 A0A182JLU4 A0A2J7PNS5 U5EJ77 A0A182Y5G8 A0A182UWJ2 A0A182U4A5 A0A182L773 A0A182XG84 Q7QB34 A0A182IDC9 A0A1Q3FDX9 G1K2H7 A0A151PE10 A0A182R0M6 W4YGM6 A0A182LV58 A0A1W5A2T8 A0A210PPX6 A0A182RZJ2 B0WU66 A0A182F7T8 G3TWS7 A0A2Y9ECN0 A0A023EMC6 A0A182NLF8 W5JLQ1 E3TEK2 A0A1B6MQC3 A0A2M4ACH7 A0A2M4BZ25 T1E7B2 A0A2M3Z7X2 H2U0D4 A0A182PLV9 G1U1H3 H3CQN7
PDB
2BME
E-value=4.73471e-79,
Score=746
Ontologies
GO
GO:0003924
GO:0005525
GO:0055037
GO:0045202
GO:0007299
GO:0043025
GO:0006897
GO:0032482
GO:0005768
GO:0016192
GO:0031982
GO:0005886
GO:0004152
GO:0044205
GO:0005743
GO:0006207
GO:0030100
GO:0004767
GO:0006886
GO:0005524
GO:0019287
GO:0005829
GO:0004163
GO:0032593
GO:0007264
GO:0016070
GO:0016620
GO:0005216
GO:0006811
GO:0016787
GO:0003676
GO:0016491
GO:0015930
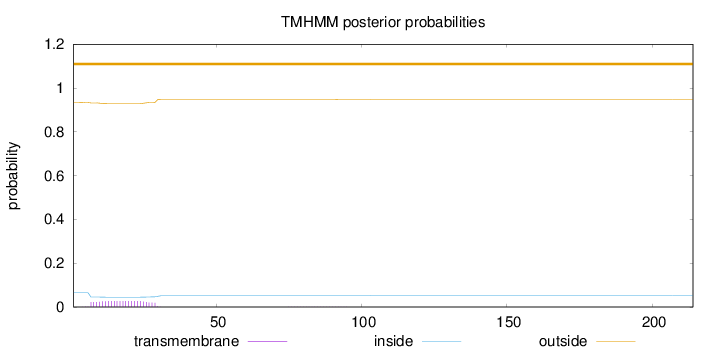
Topology
Subcellular location
Mitochondrion inner membrane
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.585819999999999
Exp number, first 60 AAs:
0.576409999999999
Total prob of N-in:
0.06524
outside
1 - 214
Population Genetic Test Statistics
Pi
175.07345
Theta
195.410038
Tajima's D
-0.367249
CLR
87.171285
CSRT
0.271636418179091
Interpretation
Uncertain
