Gene
KWMTBOMO13772
Pre Gene Modal
BGIBMGA011478
Annotation
PREDICTED:_Hermansky-Pudlak_syndrome_1_protein_homolog_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.635 Nuclear Reliability : 1.168
Sequence
CDS
ATGCAGATAAAGAGCACGGTGCGGCGAGCGCTGAGCGTGCTGCGCGAGGGGTACAGCGGGGCGACGTGGCGGCGCGGCGCGTTCCACGTGTGCGCGGTGCGGTGGTGGCAGCGCGGCGCCGCGCCCTGCCGCCCCGCGCGCCCCCCGCCGCCCGCCGCCCTGCGCGCGCTGCCGCCGCCCGGGGACATCCTCGCCTCGTTCTACAGGCAATTGCTCGAGCAGTCATTTCCAAACGACAGCCAGGGAGTGAACATCAAGGAGTTGATATGCGTCCACTTGGGCCTGTTGCCGGCTTCGACGGCCGTGCAGCAGGCGCGCCGCCTCGCGCACTCCGTACACGAACTCGCCAGCGACATCCCCGCCATCGCCGCAGACCTACTCTGA
Protein
MQIKSTVRRALSVLREGYSGATWRRGAFHVCAVRWWQRGAAPCRPARPPPPAALRALPPPGDILASFYRQLLEQSFPNDSQGVNIKELICVHLGLLPASTAVQQARRLAHSVHELASDIPAIAADLL
Summary
Uniprot
A0A3S2N9P9
A0A194RUM4
A0A0N0PA96
A0A212FE43
A0A0L7LKZ6
E0VH59
+ More
A0A2A3EG60 A0A2J7RH89 A0A3R7PTJ2 A0A2J7RH88 A0A067R1G5 A0A0L7QPW2 A0A154PKQ3 K7INS4 A0A232FCZ0 A0A1B6KVR4 A0A226ERW5 A0A1B6LXC4 A0A1B6D2N5 A0A0M9AAT9 A0A1B6HX39 A0A034VAH9 B4MY78 A0A0A1X2D0 A0A1Y1JVM4 A0A1Y1JYQ2 A0A023F1I4 A0A0V0GDC6 B3MC75 E9J727 E2C4M1 A0A151WEZ8 A0A224XGS9 A0A1W4UHQ5 E2AB18 A0A158NZG2 A0A2P2I9I4 J9K8N1 A0A2S2PYT2 A0A2S2NX13 B3NQR4 A0A3Q1H0U6 A0A0A9VYF7 A0A146LGN4 A0A1A8EPC4 B4P7Q4 A0A3B4TYC0 A0A3B4Y9N4 Q7K3N1 F6WUZ3 A0A0J9U278 B4QGA6 A0A1A8MXC4 D6WAY1 A0A2M4APH8 A0A1B6DMF9 B4JW80 B4HRQ7 A0A0T6AWE7 A0A2M4CKP2 A0A0S7KZE1 A0A3Q0T2W6 G3NXA1 A0A3B0JM72 A0A3Q2YNM6 A0A3P9LQ36 H2LUH3 A0A3P9J2I7 A0A2U9CE79 A0A3Q3WAG5 A0A3P9Q6E3 T1DGI6 A0A3Q3GGA4 A0A3Q0JDU0 L5MBM0 S7NPJ2 A0A096MD29 A0A1D2MVD0 B0XEH2 A0A1A8CQ28 A0A336LSK3 A0A1A7WLY1 A0A1A7ZFG4 A0A1A8NJ24 A0A0L0C8R6
A0A2A3EG60 A0A2J7RH89 A0A3R7PTJ2 A0A2J7RH88 A0A067R1G5 A0A0L7QPW2 A0A154PKQ3 K7INS4 A0A232FCZ0 A0A1B6KVR4 A0A226ERW5 A0A1B6LXC4 A0A1B6D2N5 A0A0M9AAT9 A0A1B6HX39 A0A034VAH9 B4MY78 A0A0A1X2D0 A0A1Y1JVM4 A0A1Y1JYQ2 A0A023F1I4 A0A0V0GDC6 B3MC75 E9J727 E2C4M1 A0A151WEZ8 A0A224XGS9 A0A1W4UHQ5 E2AB18 A0A158NZG2 A0A2P2I9I4 J9K8N1 A0A2S2PYT2 A0A2S2NX13 B3NQR4 A0A3Q1H0U6 A0A0A9VYF7 A0A146LGN4 A0A1A8EPC4 B4P7Q4 A0A3B4TYC0 A0A3B4Y9N4 Q7K3N1 F6WUZ3 A0A0J9U278 B4QGA6 A0A1A8MXC4 D6WAY1 A0A2M4APH8 A0A1B6DMF9 B4JW80 B4HRQ7 A0A0T6AWE7 A0A2M4CKP2 A0A0S7KZE1 A0A3Q0T2W6 G3NXA1 A0A3B0JM72 A0A3Q2YNM6 A0A3P9LQ36 H2LUH3 A0A3P9J2I7 A0A2U9CE79 A0A3Q3WAG5 A0A3P9Q6E3 T1DGI6 A0A3Q3GGA4 A0A3Q0JDU0 L5MBM0 S7NPJ2 A0A096MD29 A0A1D2MVD0 B0XEH2 A0A1A8CQ28 A0A336LSK3 A0A1A7WLY1 A0A1A7ZFG4 A0A1A8NJ24 A0A0L0C8R6
Pubmed
26354079
22118469
26227816
20566863
24845553
20075255
+ More
28648823 25348373 17994087 25830018 28004739 25474469 21282665 20798317 21347285 25401762 26823975 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19892987 22936249 18362917 19820115 17554307 27289101 26108605
28648823 25348373 17994087 25830018 28004739 25474469 21282665 20798317 21347285 25401762 26823975 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19892987 22936249 18362917 19820115 17554307 27289101 26108605
EMBL
RSAL01000399
RVE41939.1
KQ459833
KPJ19841.1
KQ459037
KPJ04274.1
+ More
AGBW02008970 OWR52029.1 JTDY01000716 KOB76112.1 DS235161 EEB12715.1 KZ288266 PBC30282.1 NEVH01003746 PNF40203.1 QCYY01000856 ROT82225.1 PNF40202.1 KK853097 KDR11428.1 KQ414799 KOC60670.1 KQ434948 KZC12432.1 NNAY01000445 OXU28328.1 GEBQ01024643 JAT15334.1 LNIX01000002 OXA60249.1 GEBQ01011650 JAT28327.1 GEDC01017425 JAS19873.1 KQ435698 KOX80690.1 GECU01028480 JAS79226.1 GAKP01020384 JAC38568.1 CH963894 EDW77067.1 GBXI01009392 JAD04900.1 GEZM01099380 JAV53359.1 GEZM01099378 JAV53361.1 GBBI01003858 JAC14854.1 GECL01000802 JAP05322.1 CH902619 EDV36175.1 GL768421 EFZ11344.1 GL452536 EFN77101.1 KQ983238 KYQ46387.1 GFTR01006192 JAW10234.1 GL438234 EFN69294.1 ADTU01004592 IACF01005082 LAB70669.1 ABLF02037943 GGMS01001484 MBY70687.1 GGMR01008889 MBY21508.1 CH954179 EDV55974.1 GBHO01042367 JAG01237.1 GDHC01012483 JAQ06146.1 HAEB01002195 SBQ48722.1 CM000158 EDW91080.1 AE013599 AY058469 AAF58194.1 AAL13698.1 CM002911 KMY93855.1 CM000362 EDX07144.1 HAEF01020118 SBR61277.1 KQ971312 EEZ98699.2 GGFK01009352 MBW42673.1 GEDC01010402 JAS26896.1 CH916375 EDV98218.1 CH480816 EDW47921.1 LJIG01022706 KRT79134.1 GGFL01001706 MBW65884.1 GBYX01191545 JAO82662.1 OUUW01000001 SPP74376.1 CP026257 AWP14009.1 GAMD01002761 JAA98829.1 KB102330 ELK35670.1 KE164716 EPQ19694.1 AYCK01014814 LJIJ01000476 ODM97047.1 DS232831 EDS25969.1 HADZ01017936 SBP81877.1 UFQT01000095 SSX19693.1 HADW01005129 SBP06529.1 HADY01002904 SBP41389.1 HAEH01002487 HAEI01014043 SBR69023.1 JRES01000755 KNC28627.1
AGBW02008970 OWR52029.1 JTDY01000716 KOB76112.1 DS235161 EEB12715.1 KZ288266 PBC30282.1 NEVH01003746 PNF40203.1 QCYY01000856 ROT82225.1 PNF40202.1 KK853097 KDR11428.1 KQ414799 KOC60670.1 KQ434948 KZC12432.1 NNAY01000445 OXU28328.1 GEBQ01024643 JAT15334.1 LNIX01000002 OXA60249.1 GEBQ01011650 JAT28327.1 GEDC01017425 JAS19873.1 KQ435698 KOX80690.1 GECU01028480 JAS79226.1 GAKP01020384 JAC38568.1 CH963894 EDW77067.1 GBXI01009392 JAD04900.1 GEZM01099380 JAV53359.1 GEZM01099378 JAV53361.1 GBBI01003858 JAC14854.1 GECL01000802 JAP05322.1 CH902619 EDV36175.1 GL768421 EFZ11344.1 GL452536 EFN77101.1 KQ983238 KYQ46387.1 GFTR01006192 JAW10234.1 GL438234 EFN69294.1 ADTU01004592 IACF01005082 LAB70669.1 ABLF02037943 GGMS01001484 MBY70687.1 GGMR01008889 MBY21508.1 CH954179 EDV55974.1 GBHO01042367 JAG01237.1 GDHC01012483 JAQ06146.1 HAEB01002195 SBQ48722.1 CM000158 EDW91080.1 AE013599 AY058469 AAF58194.1 AAL13698.1 CM002911 KMY93855.1 CM000362 EDX07144.1 HAEF01020118 SBR61277.1 KQ971312 EEZ98699.2 GGFK01009352 MBW42673.1 GEDC01010402 JAS26896.1 CH916375 EDV98218.1 CH480816 EDW47921.1 LJIG01022706 KRT79134.1 GGFL01001706 MBW65884.1 GBYX01191545 JAO82662.1 OUUW01000001 SPP74376.1 CP026257 AWP14009.1 GAMD01002761 JAA98829.1 KB102330 ELK35670.1 KE164716 EPQ19694.1 AYCK01014814 LJIJ01000476 ODM97047.1 DS232831 EDS25969.1 HADZ01017936 SBP81877.1 UFQT01000095 SSX19693.1 HADW01005129 SBP06529.1 HADY01002904 SBP41389.1 HAEH01002487 HAEI01014043 SBR69023.1 JRES01000755 KNC28627.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000037510
UP000009046
+ More
UP000242457 UP000235965 UP000283509 UP000027135 UP000053825 UP000076502 UP000002358 UP000215335 UP000198287 UP000053105 UP000007798 UP000007801 UP000008237 UP000075809 UP000192221 UP000000311 UP000005205 UP000007819 UP000008711 UP000265040 UP000002282 UP000261420 UP000261360 UP000000803 UP000002281 UP000000304 UP000007266 UP000001070 UP000001292 UP000261340 UP000007635 UP000268350 UP000264820 UP000265180 UP000001038 UP000265200 UP000246464 UP000261620 UP000242638 UP000261660 UP000079169 UP000028760 UP000094527 UP000002320 UP000037069
UP000242457 UP000235965 UP000283509 UP000027135 UP000053825 UP000076502 UP000002358 UP000215335 UP000198287 UP000053105 UP000007798 UP000007801 UP000008237 UP000075809 UP000192221 UP000000311 UP000005205 UP000007819 UP000008711 UP000265040 UP000002282 UP000261420 UP000261360 UP000000803 UP000002281 UP000000304 UP000007266 UP000001070 UP000001292 UP000261340 UP000007635 UP000268350 UP000264820 UP000265180 UP000001038 UP000265200 UP000246464 UP000261620 UP000242638 UP000261660 UP000079169 UP000028760 UP000094527 UP000002320 UP000037069
Interpro
IPR026053
HPS1
ProteinModelPortal
A0A3S2N9P9
A0A194RUM4
A0A0N0PA96
A0A212FE43
A0A0L7LKZ6
E0VH59
+ More
A0A2A3EG60 A0A2J7RH89 A0A3R7PTJ2 A0A2J7RH88 A0A067R1G5 A0A0L7QPW2 A0A154PKQ3 K7INS4 A0A232FCZ0 A0A1B6KVR4 A0A226ERW5 A0A1B6LXC4 A0A1B6D2N5 A0A0M9AAT9 A0A1B6HX39 A0A034VAH9 B4MY78 A0A0A1X2D0 A0A1Y1JVM4 A0A1Y1JYQ2 A0A023F1I4 A0A0V0GDC6 B3MC75 E9J727 E2C4M1 A0A151WEZ8 A0A224XGS9 A0A1W4UHQ5 E2AB18 A0A158NZG2 A0A2P2I9I4 J9K8N1 A0A2S2PYT2 A0A2S2NX13 B3NQR4 A0A3Q1H0U6 A0A0A9VYF7 A0A146LGN4 A0A1A8EPC4 B4P7Q4 A0A3B4TYC0 A0A3B4Y9N4 Q7K3N1 F6WUZ3 A0A0J9U278 B4QGA6 A0A1A8MXC4 D6WAY1 A0A2M4APH8 A0A1B6DMF9 B4JW80 B4HRQ7 A0A0T6AWE7 A0A2M4CKP2 A0A0S7KZE1 A0A3Q0T2W6 G3NXA1 A0A3B0JM72 A0A3Q2YNM6 A0A3P9LQ36 H2LUH3 A0A3P9J2I7 A0A2U9CE79 A0A3Q3WAG5 A0A3P9Q6E3 T1DGI6 A0A3Q3GGA4 A0A3Q0JDU0 L5MBM0 S7NPJ2 A0A096MD29 A0A1D2MVD0 B0XEH2 A0A1A8CQ28 A0A336LSK3 A0A1A7WLY1 A0A1A7ZFG4 A0A1A8NJ24 A0A0L0C8R6
A0A2A3EG60 A0A2J7RH89 A0A3R7PTJ2 A0A2J7RH88 A0A067R1G5 A0A0L7QPW2 A0A154PKQ3 K7INS4 A0A232FCZ0 A0A1B6KVR4 A0A226ERW5 A0A1B6LXC4 A0A1B6D2N5 A0A0M9AAT9 A0A1B6HX39 A0A034VAH9 B4MY78 A0A0A1X2D0 A0A1Y1JVM4 A0A1Y1JYQ2 A0A023F1I4 A0A0V0GDC6 B3MC75 E9J727 E2C4M1 A0A151WEZ8 A0A224XGS9 A0A1W4UHQ5 E2AB18 A0A158NZG2 A0A2P2I9I4 J9K8N1 A0A2S2PYT2 A0A2S2NX13 B3NQR4 A0A3Q1H0U6 A0A0A9VYF7 A0A146LGN4 A0A1A8EPC4 B4P7Q4 A0A3B4TYC0 A0A3B4Y9N4 Q7K3N1 F6WUZ3 A0A0J9U278 B4QGA6 A0A1A8MXC4 D6WAY1 A0A2M4APH8 A0A1B6DMF9 B4JW80 B4HRQ7 A0A0T6AWE7 A0A2M4CKP2 A0A0S7KZE1 A0A3Q0T2W6 G3NXA1 A0A3B0JM72 A0A3Q2YNM6 A0A3P9LQ36 H2LUH3 A0A3P9J2I7 A0A2U9CE79 A0A3Q3WAG5 A0A3P9Q6E3 T1DGI6 A0A3Q3GGA4 A0A3Q0JDU0 L5MBM0 S7NPJ2 A0A096MD29 A0A1D2MVD0 B0XEH2 A0A1A8CQ28 A0A336LSK3 A0A1A7WLY1 A0A1A7ZFG4 A0A1A8NJ24 A0A0L0C8R6
Ontologies
PANTHER
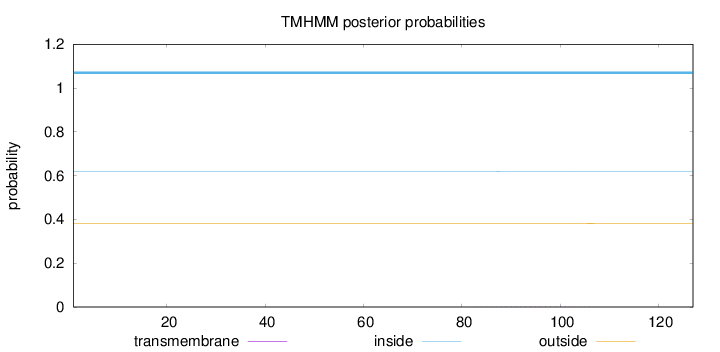
Topology
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0258
Exp number, first 60 AAs:
0.00142
Total prob of N-in:
0.62012
inside
1 - 127
Population Genetic Test Statistics
Pi
73.340825
Theta
110.211974
Tajima's D
-0.522516
CLR
50.311838
CSRT
0.239838008099595
Interpretation
Uncertain
